Toxicogenomic profiling of chemically exposed humans in risk assessment
- PMID: 20382258
- PMCID: PMC2928857
- DOI: 10.1016/j.mrrev.2010.04.001
Toxicogenomic profiling of chemically exposed humans in risk assessment
Abstract
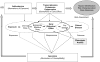
Gene-environment interactions contribute to complex disease development. The environmental contribution, in particular low-level and prevalent environmental exposures, may constitute much of the risk and contribute substantially to disease. Systematic risk evaluation of the majority of human chemical exposures, has not been conducted and is a goal of regulatory agencies in the U.S. and worldwide. With the recent recognition that toxicological approaches more predictive of effects in humans are required for risk assessment, in vitro human cell line data as well as animal data are being used to identify toxicity mechanisms that can be translated into biomarkers relevant to human exposure studies. In this review, we discuss how data from toxicogenomic studies of exposed human populations can inform risk assessment, by generating biomarkers of exposure, early effect, and/or susceptibility, elucidating mechanisms of action underlying exposure-related disease, and detecting response at low doses. Good experimental design incorporating precise, individual exposure measurements, phenotypic anchors (pre-disease or traditional toxicological markers), and a range of relevant exposure levels, is necessary. Further, toxicogenomic studies need to be designed with sufficient power to detect true effects of the exposure. As more studies are performed and incorporated into databases such as the Comparative Toxicogenomics Database (CTD) and Chemical Effects in Biological Systems (CEBS), data can be mined for classification of newly tested chemicals (hazard identification), and, for investigating the dose-response, and inter-relationship among genes, environment and disease in a systems biology approach (risk characterization).
Copyright © 2010 Elsevier B.V. All rights reserved.
Conflict of interest statement
Dr. Smith has received consulting and expert testimony fees from lawyers representing both plaintiffs and defendants in cases involving claims related to exposure to benzene.
Figures
References
-
- Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847–1850. - PubMed
-
- Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk of complex disease. Curr Opin Genet Dev. 2008;18:257–263. - PubMed
-
- McCarroll SA. Extending genome-wide association studies to copy-number variation. Hum Mol Genet. 2008;17:R135–142. - PubMed
-
- Robertson TL, Kato H, Gordon T, Kagan A, Rhoads GG, Land CE, Worth RM, Belsky JL, Dock DS, Miyanishi M, Kawamoto S. Epidemiologic studies of coronary heart disease and stroke in Japanese men living in Japan, Hawaii and California. Coronary heart disease risk factors in Japan and Hawaii. Am J Cardiol. 1977;39:244–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

