Single-molecule force spectroscopy approach to enzyme catalysis
- PMID: 20382731
- PMCID: PMC2885171
- DOI: 10.1074/jbc.R109.011932
Single-molecule force spectroscopy approach to enzyme catalysis
Abstract
Enzyme catalysis has been traditionally studied using a diverse set of techniques such as bulk biochemistry, x-ray crystallography, and NMR. Recently, single-molecule force spectroscopy by atomic force microscopy has been used as a new tool to study the catalytic properties of an enzyme. In this approach, a mechanical force ranging up to hundreds of piconewtons is applied to the substrate of an enzymatic reaction, altering the conformational energy of the substrate-enzyme interactions during catalysis. From these measurements, the force dependence of an enzymatic reaction can be determined. The force dependence provides valuable new information about the dynamics of enzyme catalysis with sub-angstrom resolution, a feat unmatched by any other current technique. To date, single-molecule force spectroscopy has been applied to gain insight into the reduction of disulfide bonds by different enzymes of the thioredoxin family. This minireview aims to present a perspective on this new approach to study enzyme catalysis and to summarize the results that have already been obtained from it. Finally, the specific requirements that must be fulfilled to apply this new methodology to any other enzyme will be discussed.
Figures
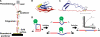

References
-
- Kraut D. A., Carroll K. S., Herschlag D. (2003) Annu. Rev. Biochem. 72, 517–571 - PubMed
-
- Benkovic S. J., Hammes-Schiffer S. (2003) Science 301, 1196–1202 - PubMed
-
- Jäckel C., Kast P., Hilvert D. (2008) Annu. Rev. Biophys. 37, 153–173 - PubMed
-
- Karplus M., McCammon J. A. (1983) Annu. Rev. Biochem. 52, 263–300 - PubMed
-
- Benkovic S. J., Hammes G. G., Hammes-Schiffer S. (2008) Biochemistry 47, 3317–3321 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

