Timing, coordination, and rhythm: acrobatics at the DNA replication fork
- PMID: 20382733
- PMCID: PMC2885174
- DOI: 10.1074/jbc.R109.022939
Timing, coordination, and rhythm: acrobatics at the DNA replication fork
Abstract
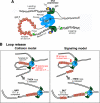
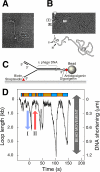
In DNA replication, the antiparallel nature of the parental duplex imposes certain constraints on the activity of the DNA polymerases that synthesize new DNA. The leading-strand polymerase advances in a continuous fashion, but the lagging-strand polymerase is forced to restart at short intervals. In several prokaryotic systems studied so far, this problem is solved by the formation of a loop in the lagging strand of the replication fork to reorient the lagging-strand DNA polymerase so that it advances in parallel with the leading-strand polymerase. The replication loop grows and shrinks during each cycle of Okazaki fragment synthesis. The timing of Okazaki fragment synthesis and loop formation is determined by a subtle interplay of enzymatic activities at the fork. Recent developments in single-molecule techniques have enabled the direct observation of these processes and have greatly contributed to a better understanding of the dynamic nature of the replication fork. Here, we will review recent experimental advances, present the current models, and discuss some of the exciting developments in the field.
Figures
References
-
- Benkovic S. J., Valentine A. M., Salinas F. (2001) Annu. Rev. Biochem. 70, 181–208 - PubMed
-
- Johnson A., O'Donnell M. (2005) Annu. Rev. Biochem. 74, 283–315 - PubMed
-
- Hamdan S. M., Richardson C. C. (2009) Annu. Rev. Biochem. 78, 205–243 - PubMed
-
- Ogawa T., Okazaki T. (1980) Annu. Rev. Biochem. 49, 421–457 - PubMed
-
- Tabor S., Huber H. E., Richardson C. C. (1987) J. Biol. Chem. 262, 16212–16223 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

