RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43
- PMID: 20382770
- PMCID: PMC2901686
- DOI: 10.1128/JB.00031-10
RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43
Abstract
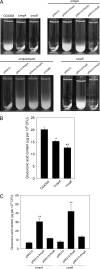
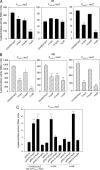
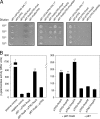
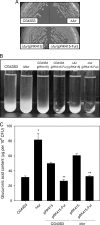
Sequence analysis of the large virulence plasmid pLVPK in Klebsiella pneumoniae CG43 revealed the presence of another mucoid factor encoding gene rmpA besides rmpA2. Promoter activity measurement indicated that the deletion of rmpA reduced K2 capsular polysaccharide (CPS) biosynthesis, resulting in decreased colony mucoidy and virulence in mice. Introduction of a multicopy plasmid carrying rmpA restored CPS production in the rmpA or rmpA2 mutant but not in the rcsB mutant. Transformation of the rmpA deletion mutant with an rcsB-carrying plasmid also failed to enhance CPS production, suggesting that a cooperation of RmpA with RcsB is required for regulatory activity. This was further corroborated by the demonstration of in vivo interaction between RmpA and RcsB using two-hybrid analysis and coimmunoprecipitation analysis. A putative Fur binding box was only found at the 5' noncoding region of rmpA. The promoter activity analysis indicated that the deletion of fur increased the rmpA promoter activity. Using electrophoretic mobility shift assay, we further demonstrated that Fur exerts its regulatory activity by binding directly to the promoter. As a result, the fur deletion mutant exhibited an increase in colony mucoidy, CPS production, and virulence in mice. In summary, our results suggested that RmpA activates CPS biosynthesis in K. pneumoniae CG43 via an RcsB-dependent manner. The expression of rmpA is regulated by the availability of iron and is negatively controlled by Fur.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

