phrR-like gene praR of Azorhizobium caulinodans ORS571 is essential for symbiosis with Sesbania rostrata and is involved in expression of reb genes
- PMID: 20382809
- PMCID: PMC2876453
- DOI: 10.1128/AEM.00238-10
phrR-like gene praR of Azorhizobium caulinodans ORS571 is essential for symbiosis with Sesbania rostrata and is involved in expression of reb genes
Abstract
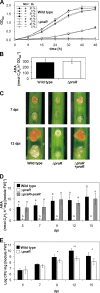
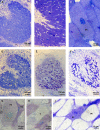
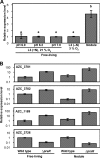
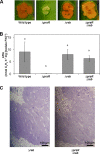
This study focuses on the function of the gene praR that encodes a putative transcription factor in Azorhizobium caulinodans ORS571, a microsymbiont of Sesbania rostrata. The praR gene is a homolog of the phrR gene of Sinorhizobium medicae WSM419, and the praR and phrR homologs are distributed throughout the class Alphaproteobacteria. The growth and nitrogen fixation activity of an A. caulinodans praR deletion mutant in the free-living state were not significantly different from those of the wild-type strain. However, the stem nodules formed by the praR mutant showed lower nitrogen fixation activity than the wild-type stem nodules. Microscopy revealed that infected host cells with an oval or elongated shape were observed at early stages in the nodules formed by the praR mutant, but these infected cells gradually fell into two types. One maintained an oval or elongated shape, but the vacuoles in these cells gradually enlarged and the bacteria gradually disappeared. The other cells were shrunken with bacteria remaining inside. Microarrays revealed that genes homologous to the reb genes of Caedibacter taeniospiralis were highly expressed in the praR mutant. Furthermore, the stem nodules formed by an A. caulinodans mutant with a deletion of praR and reb-homologous genes showed high nitrogen fixation activity, comparable to that of the wild-type stem nodules, and were filled with oval or elongated host cells. These results suggest that PraR controls the expression of the reb-homologous genes and that high expression of reb-homologous genes causes aberrance in A. caulinodans-S. rostrata symbiosis.
Figures

References
-
- Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. - PubMed
-
- Beier, C. L., M. Horn, R. Michel, M. Schweikert, H. Görtz, and M. Wagner. 2002. The genus Caedibacter comprises endosymbionts of Paramecium spp. related to the Rickettsiales (Alphaproteobacteria) and to Francisella tularensis (Gammaproteobacteria). Appl. Environ. Microbiol. 68:6043-6050. - PMC - PubMed
-
- Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. - PubMed
-
- Bittel, P., and S. Robatzek. 2007. Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr. Opin. Plant Biol. 10:335-341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

