Experimental phasing with SHELXC/D/E: combining chain tracing with density modification
- PMID: 20383001
- PMCID: PMC2852312
- DOI: 10.1107/S0907444909038360
Experimental phasing with SHELXC/D/E: combining chain tracing with density modification
Abstract
The programs SHELXC, SHELXD and SHELXE are designed to provide simple, robust and efficient experimental phasing of macromolecules by the SAD, MAD, SIR, SIRAS and RIP methods and are particularly suitable for use in automated structure-solution pipelines. This paper gives a general account of experimental phasing using these programs and describes the extension of iterative density modification in SHELXE by the inclusion of automated protein main-chain tracing. This gives a good indication as to whether the structure has been solved and enables interpretable maps to be obtained from poorer starting phases. The autotracing algorithm starts with the location of possible seven-residue alpha-helices and common tripeptides. After extension of these fragments in both directions, various criteria are used to decide whether to accept or reject the resulting poly-Ala traces. Noncrystallographic symmetry (NCS) is applied to the traced fragments, not to the density. Further features are the use of a 'no-go' map to prevent the traces from passing through heavy atoms or symmetry elements and a splicing technique to combine the best parts of traces (including those generated by NCS) that partly overlap.
Figures

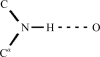



References
-
- Abrahams, J. P. (1997). Acta Cryst. D53, 371–376. - PubMed
-
- Bricogne, G. (1976). Acta Cryst. A32, 832–847.
-
- Caliandro, R., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C. & Siliqi, D. (2005). Acta Cryst. D61, 556–565. - PubMed
-
- Cowtan, K. (2000). Acta Cryst. D56, 1612–1621. - PubMed
-
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

