Features and development of Coot
- PMID: 20383002
- PMCID: PMC2852313
- DOI: 10.1107/S0907444910007493
Features and development of Coot
Abstract
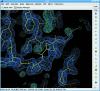
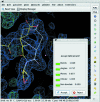
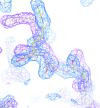
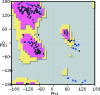
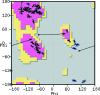
Coot is a molecular-graphics application for model building and validation of biological macromolecules. The program displays electron-density maps and atomic models and allows model manipulations such as idealization, real-space refinement, manual rotation/translation, rigid-body fitting, ligand search, solvation, mutations, rotamers and Ramachandran idealization. Furthermore, tools are provided for model validation as well as interfaces to external programs for refinement, validation and graphics. The software is designed to be easy to learn for novice users, which is achieved by ensuring that tools for common tasks are 'discoverable' through familiar user-interface elements (menus and toolbars) or by intuitive behaviour (mouse controls). Recent developments have focused on providing tools for expert users, with customisable key bindings, extensions and an extensive scripting interface. The software is under rapid development, but has already achieved very widespread use within the crystallographic community. The current state of the software is presented, with a description of the facilities available and of some of the underlying methods employed.
Figures

References
-
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. - PubMed
-
- Bernstein, F. C., Koetzle, T. F., Williams, G. J. B., Meyer, E. F. Jr, Brice, M. D., Rodgers, J. R., Kennard, O., Shimanouchi, T. & Tasumi, M. (1977). J. Mol. Biol.112, 535–542. - PubMed
-
- Blanc, E., Roversi, P., Vonrhein, C., Flensburg, C., Lea, S. M. & Bricogne, G. (2004). Acta Cryst. D60, 2210–2221. - PubMed
-
- Blundell, T. L., Jhoti, H. & Abell, C. (2002). Nature Rev. Drug Discov.1, 45–54. - PubMed
-
- Brünger, A. T. (1992). X-PLOR v.3.1. A System for X-ray Crystallography and NMR. New Haven: Yale University Press.
Publication types
MeSH terms
Substances
Grants and funding
- BB/D522403/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F020368/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 GM087721/GM/NIGMS NIH HHS/United States
- BB/D522403/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBF0202281/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

