Structures of influenza A proteins and insights into antiviral drug targets
- PMID: 20383144
- PMCID: PMC2957899
- DOI: 10.1038/nsmb.1779
Structures of influenza A proteins and insights into antiviral drug targets
Abstract
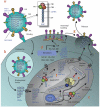
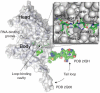
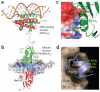
The world is currently undergoing a pandemic caused by an H1N1 influenza A virus, the so-called 'swine flu'. The H5N1 ('bird flu') influenza A viruses, now circulating in Asia, Africa and Europe, are extremely virulent in humans, although they have not so far acquired the ability to transfer efficiently from human to human. These health concerns have spurred considerable interest in understanding the molecular biology of influenza A viruses. Recent structural studies of influenza A virus proteins (or fragments) help enhance our understanding of the molecular mechanisms of the viral proteins and the effects of drug resistance to improve drug design. The structures of domains of the influenza RNA-dependent RNA polymerase and the nonstructural NS1A protein provide opportunities for targeting these proteins to inhibit viral replication.
Figures




References
-
- Reid AH, Taubenberger JK, Fanning TG. The 1918 Spanish influenza: integrating history and biology. Microbes Infect. 2001;3:81–87. - PubMed
-
- Chowell G, et al. Severe respiratory disease concurrent with the circulation of H1N1 influenza. N. Engl. J. Med. 2009;361:674–679. - PubMed
-
- Lamb RA, Krug RM. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 4th edn. Lippincott, Williams & Wilkins; Philadelphia, USA: 2001. pp. 1487–1532.
-
- Wright PF, Webster RG. Orthomyxoviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 4th edn. Lippincott, Williams & Wilkins; Philadelphia, USA: 2001. pp. 1533–1579.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

