High-throughput computational structure-based characterization of protein families: START domains and implications for structural genomics
- PMID: 20383749
- PMCID: PMC2881152
- DOI: 10.1007/s10969-010-9086-7
High-throughput computational structure-based characterization of protein families: START domains and implications for structural genomics
Abstract
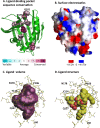
SkyLine, a high-throughput homology modeling pipeline tool, detects and models true sequence homologs to a given protein structure. Structures and models are stored in SkyBase with links to computational function annotation, as calculated by MarkUs. The SkyLine/SkyBase/MarkUs technology represents a novel structure-based approach that is more objective and versatile than other protein classification resources. This structure-centric strategy provides a multi-dimensional organization and coverage of protein space at the levels of family, function, and genome. The concept of "modelability", the ability to model sequences on related structures, provides a reliable criterion for membership in a protein family ("leverage") and underlies the unique success of this approach. The overall procedure is illustrated by its application to START domains, which comprise a Biomedical Theme for the Northeast Structural Genomics Consortium as part of the Protein Structure Initiative. START domains are typically involved in the non-vesicular transport of lipids. While 19 experimentally determined structures are available, the family, whose evolutionary hierarchy is not well determined, is highly sequence diverse, and the ligand-binding potential of many family members is unknown. The SkyLine/SkyBase/MarkUs approach provides significant insights and predicts: (1) many more family members (approximately 4,000) than any other resource; (2) the function for a large number of unannotated proteins; (3) instances of START domains in genomes from which they were thought to be absent; and (4) the existence of two types of novel proteins, those containing dual START domain and those containing N-terminal START domains.
Figures



Similar articles
-
Strategies for high-throughput comparative modeling: applications to leverage analysis in structural genomics and protein family organization.Proteins. 2007 Mar 1;66(4):766-77. doi: 10.1002/prot.21191. Proteins. 2007. PMID: 17154423
-
NMR in structural genomics to increase structural coverage of the protein universe: Delivered by Prof. Kurt Wüthrich on 7 July 2013 at the 38th FEBS Congress in St. Petersburg, Russia.FEBS J. 2016 Nov;283(21):3870-3881. doi: 10.1111/febs.13751. Epub 2016 Jun 9. FEBS J. 2016. PMID: 27154589 Free PMC article.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
A domain-centric solution to functional genomics via dcGO Predictor.BMC Bioinformatics. 2013;14 Suppl 3(Suppl 3):S9. doi: 10.1186/1471-2105-14-S3-S9. Epub 2013 Feb 28. BMC Bioinformatics. 2013. PMID: 23514627 Free PMC article.
-
PSI-2: structural genomics to cover protein domain family space.Structure. 2009 Jun 10;17(6):869-81. doi: 10.1016/j.str.2009.03.015. Structure. 2009. PMID: 19523904 Free PMC article. Review.
Cited by
-
The Lipid Transfer Protein StarD7: Structure, Function, and Regulation.Int J Mol Sci. 2013 Mar 18;14(3):6170-86. doi: 10.3390/ijms14036170. Int J Mol Sci. 2013. PMID: 23507753 Free PMC article.
-
Genome-wide structural analysis reveals novel membrane binding properties of AP180 N-terminal homology (ANTH) domains.J Biol Chem. 2011 Sep 30;286(39):34155-63. doi: 10.1074/jbc.M111.265611. Epub 2011 Aug 2. J Biol Chem. 2011. PMID: 21828048 Free PMC article.
-
Using structure to explore the sequence alignment space of remote homologs.PLoS Comput Biol. 2011 Oct;7(10):e1002175. doi: 10.1371/journal.pcbi.1002175. Epub 2011 Oct 6. PLoS Comput Biol. 2011. PMID: 21998567 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

