KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans
- PMID: 20384682
- PMCID: PMC2969852
- DOI: 10.1111/j.1365-2958.2010.07119.x
KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans
Abstract
The polysaccharide beta-1,6-glucan is a major component of the cell wall of Cryptococcus neoformans, but its function has not been investigated in this fungal pathogen. We have identified and characterized seven genes, belonging to the KRE family, which are putatively involved in beta-1,6-glucan synthesis. The H99 deletion mutants kre5Delta and kre6Deltaskn1Delta contained less cell wall beta-1,6-glucan, grew slowly with an aberrant morphology, were highly sensitive to environmental and chemical stress and were avirulent in a mouse inhalation model of infection. These two mutants displayed alterations in cell wall chitosan and the exopolysaccharide capsule, a primary cryptococcal virulence determinant. The cell wall content of the GPI-anchored phospholipase B1 (Plb1) enzyme, which is required for cryptococcal cell wall integrity and virulence, was reduced in kre5Delta and kre6Deltaskn1Delta. Our results indicate that KRE5, KRE6 and SKN1 are involved in beta-1,6-glucan synthesis, maintenance of cell wall integrity and retention of mannoproteins and known cryptococcal virulence factors in the cell wall of C. neoformans. This study sets the stage for future investigations into the function of this abundant cell wall polymer.
Figures









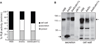
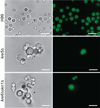
Similar articles
-
Aspartyl peptidase May1 induces host inflammatory response by altering cell wall composition in the fungal pathogen Cryptococcus neoformans.mBio. 2024 Jun 12;15(6):e0092024. doi: 10.1128/mbio.00920-24. Epub 2024 May 14. mBio. 2024. PMID: 38742885 Free PMC article.
-
Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity.J Biol Chem. 2007 Dec 28;282(52):37508-14. doi: 10.1074/jbc.M707913200. Epub 2007 Oct 18. J Biol Chem. 2007. PMID: 17947228
-
UDP-Glucuronic Acid Transport Is Required for Virulence of Cryptococcus neoformans.mBio. 2018 Jan 30;9(1):e02319-17. doi: 10.1128/mBio.02319-17. mBio. 2018. PMID: 29382737 Free PMC article.
-
Emerging themes in cryptococcal capsule synthesis.Curr Opin Struct Biol. 2011 Oct;21(5):597-602. doi: 10.1016/j.sbi.2011.08.006. Epub 2011 Sep 1. Curr Opin Struct Biol. 2011. PMID: 21889889 Free PMC article. Review.
-
The Overlooked Glycan Components of the Cryptococcus Capsule.Curr Top Microbiol Immunol. 2019;422:31-43. doi: 10.1007/82_2018_140. Curr Top Microbiol Immunol. 2019. PMID: 30203395 Review.
Cited by
-
Fungal Pathogenesis-Related Cell Wall Biogenesis, with Emphasis on the Maize Anthracnose Fungus Colletotrichum graminicola.Plants (Basel). 2022 Mar 23;11(7):849. doi: 10.3390/plants11070849. Plants (Basel). 2022. PMID: 35406829 Free PMC article. Review.
-
Peeling the onion: the outer layers of Cryptococcus neoformans.Mem Inst Oswaldo Cruz. 2018;113(7):e180040. doi: 10.1590/0074-02760180040. Epub 2018 May 7. Mem Inst Oswaldo Cruz. 2018. PMID: 29742198 Free PMC article. Review.
-
A mutated B cell chronic lymphocytic leukemia subset that recognizes and responds to fungi.J Exp Med. 2013 Jan 14;210(1):59-70. doi: 10.1084/jem.20121801. Epub 2013 Jan 7. J Exp Med. 2013. PMID: 23296468 Free PMC article.
-
Cell wall composition in Cryptococcus neoformans is media dependent and alters host response, inducing protective immunity.Front Fungal Biol. 2023;4:1183291. doi: 10.3389/ffunb.2023.1183291. Epub 2023 May 12. Front Fungal Biol. 2023. PMID: 37538303 Free PMC article.
-
Ccr4 promotes resolution of the endoplasmic reticulum stress response during host temperature adaptation in Cryptococcus neoformans.Eukaryot Cell. 2011 Jul;10(7):895-901. doi: 10.1128/EC.00006-11. Epub 2011 May 20. Eukaryot Cell. 2011. PMID: 21602483 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

