FLU, an amino acid substitution model for influenza proteins
- PMID: 20384985
- PMCID: PMC2873421
- DOI: 10.1186/1471-2148-10-99
FLU, an amino acid substitution model for influenza proteins
Abstract
Background: The amino acid substitution model is the core component of many protein analysis systems such as sequence similarity search, sequence alignment, and phylogenetic inference. Although several general amino acid substitution models have been estimated from large and diverse protein databases, they remain inappropriate for analyzing specific species, e.g., viruses. Emerging epidemics of influenza viruses raise the need for comprehensive studies of these dangerous viruses. We propose an influenza-specific amino acid substitution model to enhance the understanding of the evolution of influenza viruses.
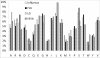
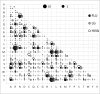
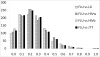
Results: A maximum likelihood approach was applied to estimate an amino acid substitution model (FLU) from approximately 113,000 influenza protein sequences, consisting of approximately 20 million residues. FLU outperforms 14 widely used models in constructing maximum likelihood phylogenetic trees for the majority of influenza protein alignments. On average, FLU gains approximately 42 log likelihood points with an alignment of 300 sites. Moreover, topologies of trees constructed using FLU and other models are frequently different. FLU does indeed have an impact on likelihood improvement as well as tree topologies. It was implemented in PhyML and can be downloaded from ftp://ftp.sanger.ac.uk/pub/1000genomes/lsq/FLU or included in PhyML 3.0 server at http://www.atgc-montpellier.fr/phyml/.
Conclusions: FLU should be useful for any influenza protein analysis system which requires an accurate description of amino acid substitutions.
Figures
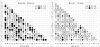
References
-
- Felsenstein J. Infering Phylogenies. Sunderland, Massachusetts, US: Sinauer Associates; 2004.
-
- Ziheng Y. Computational Molecular Evolution. 1. Oxford, UK: Oxford University Press; 2006.
-
- Opperdoes FR. In: The Phylogenetics Handbook A Practical Approach to DNA and Protein Phylogeny. Salemi M, Vandamme AM, editor. Cambridge: Cambridge University Press; 2003. Phylogenetic analysis using protein sequences; pp. 207–235.
-
- Setubal C, Meidanis J. Introduction to Computational Molecular Biology. 1. Boston, Massachusetts, US: PWS Publishing; 1997.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

