Thioredoxin and glutathione systems differ in parasitic and free-living platyhelminths
- PMID: 20385027
- PMCID: PMC2873472
- DOI: 10.1186/1471-2164-11-237
Thioredoxin and glutathione systems differ in parasitic and free-living platyhelminths
Abstract
Background: The thioredoxin and/or glutathione pathways occur in all organisms. They provide electrons for deoxyribonucleotide synthesis, function as antioxidant defenses, in detoxification, Fe/S biogenesis and participate in a variety of cellular processes. In contrast to their mammalian hosts, platyhelminth (flatworm) parasites studied so far, lack conventional thioredoxin and glutathione systems. Instead, they possess a linked thioredoxin-glutathione system with the selenocysteine-containing enzyme thioredoxin glutathione reductase (TGR) as the single redox hub that controls the overall redox homeostasis. TGR has been recently validated as a drug target for schistosomiasis and new drug leads targeting TGR have recently been identified for these platyhelminth infections that affect more than 200 million people and for which a single drug is currently available. Little is known regarding the genomic structure of flatworm TGRs, the expression of TGR variants and whether the absence of conventional thioredoxin and glutathione systems is a signature of the entire platyhelminth phylum.
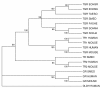
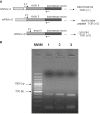
Results: We examine platyhelminth genomes and transcriptomes and find that all platyhelminth parasites (from classes Cestoda and Trematoda) conform to a biochemical scenario involving, exclusively, a selenium-dependent linked thioredoxin-glutathione system having TGR as a central redox hub. In contrast, the free-living platyhelminth Schmidtea mediterranea (Class Turbellaria) possesses conventional and linked thioredoxin and glutathione systems. We identify TGR variants in Schistosoma spp. derived from a single gene, and demonstrate their expression. We also provide experimental evidence that alternative initiation of transcription and alternative transcript processing contribute to the generation of TGR variants in platyhelminth parasites.
Conclusions: Our results indicate that thioredoxin and glutathione pathways differ in parasitic and free-living flatworms and that canonical enzymes were specifically lost in the parasitic lineage. Platyhelminth parasites possess a unique and simplified redox system for diverse essential processes, and thus TGR is an excellent drug target for platyhelminth infections. Inhibition of the central redox wire hub would lead to overall disruption of redox homeostasis and disable DNA synthesis.
Figures



Similar articles
-
Thioredoxin glutathione reductase-dependent redox networks in platyhelminth parasites.Antioxid Redox Signal. 2013 Sep 1;19(7):735-45. doi: 10.1089/ars.2012.4670. Epub 2012 Oct 3. Antioxid Redox Signal. 2013. PMID: 22909029 Free PMC article. Review.
-
Linked thioredoxin-glutathione systems in platyhelminth parasites: alternative pathways for glutathione reduction and deglutathionylation.J Biol Chem. 2011 Feb 18;286(7):4959-67. doi: 10.1074/jbc.M110.170761. Epub 2010 Nov 4. J Biol Chem. 2011. PMID: 21051543 Free PMC article.
-
Platyhelminth mitochondrial and cytosolic redox homeostasis is controlled by a single thioredoxin glutathione reductase and dependent on selenium and glutathione.J Biol Chem. 2008 Jun 27;283(26):17898-907. doi: 10.1074/jbc.M710609200. Epub 2008 Apr 11. J Biol Chem. 2008. PMID: 18408002 Free PMC article.
-
Linked thioredoxin-glutathione systems in platyhelminths.Trends Parasitol. 2004 Jul;20(7):340-6. doi: 10.1016/j.pt.2004.05.002. Trends Parasitol. 2004. PMID: 15193566 Review.
-
Thioredoxin glutathione reductase as a novel drug target: evidence from Schistosoma japonicum.PLoS One. 2012;7(2):e31456. doi: 10.1371/journal.pone.0031456. Epub 2012 Feb 22. PLoS One. 2012. PMID: 22384025 Free PMC article.
Cited by
-
Expression, crystallization and preliminary X-ray diffraction analysis of thioredoxin glutathione reductase from Schistosoma japonicum in complex with FAD.Acta Crystallogr F Struct Biol Commun. 2014 Jan;70(Pt 1):92-6. doi: 10.1107/S2053230X1303313X. Epub 2013 Dec 24. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 24419626 Free PMC article.
-
The Enzymatic and Structural Basis for Inhibition of Echinococcus granulosus Thioredoxin Glutathione Reductase by Gold(I).Antioxid Redox Signal. 2017 Dec 20;27(18):1491-1504. doi: 10.1089/ars.2016.6816. Epub 2017 Jun 26. Antioxid Redox Signal. 2017. PMID: 28463568 Free PMC article.
-
Antiparasitic Effects of Selected Isoflavones on Flatworms.Helminthologia. 2021 Feb 10;58(1):1-16. doi: 10.2478/helm-2021-0004. eCollection 2021 Mar. Helminthologia. 2021. PMID: 33664614 Free PMC article.
-
Thioredoxin glutathione reductase-dependent redox networks in platyhelminth parasites.Antioxid Redox Signal. 2013 Sep 1;19(7):735-45. doi: 10.1089/ars.2012.4670. Epub 2012 Oct 3. Antioxid Redox Signal. 2013. PMID: 22909029 Free PMC article. Review.
-
G6PDH as a key immunometabolic and redox trigger in arthropods.Front Physiol. 2023 Nov 17;14:1287090. doi: 10.3389/fphys.2023.1287090. eCollection 2023. Front Physiol. 2023. PMID: 38046951 Free PMC article. Review.
References
-
- Littlewood DTJ. In: Parasitic flatworms: molecular biology, biochemistry, immunology and physiology. 1. Maule AG, Marks NJ, editor. CAB International; The Evolution of Parasitism in Flatworms; pp. 1–36.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

