Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development
- PMID: 20385573
- PMCID: PMC2926599
- DOI: 10.1093/nar/gkq229
Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development
Abstract
Recent studies showed that small interfering RNAs (siRNAs) and Piwi-interacting RNA (piRNA) in mammalian germ cells play important roles in retrotransposon silencing and gametogenesis. However, subsequent contribution of those small RNAs to early mammalian development remains poorly understood. We investigated the expression profiles of small RNAs in mouse metaphase II oocytes, 8-16-cell stage embryos, blastocysts and the pluripotent inner cell mass (ICM) using high-throughput pyrosequencing. Here, we show that during pre-implantation development a major small RNA class changes from retrotransposon-derived small RNAs containing siRNAs and piRNAs to zygotically synthesized microRNAs (miRNAs). Some siRNAs and piRNAs are transiently upregulated and directed against specific retrotransposon classes. We also identified miRNAs expression profiles characteristic of the ICM and trophectoderm (TE) cells. Taken together, our current study reveals a major reprogramming of functional small RNAs during early mouse development from oocyte to blastocyst.
Figures
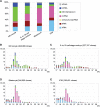

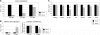


References
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
-
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. - PubMed
-
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. - PubMed

