Quantitative design and experimental validation for a single-molecule DNA nanodevice transformable among three structural states
- PMID: 20385575
- PMCID: PMC2910065
- DOI: 10.1093/nar/gkq250
Quantitative design and experimental validation for a single-molecule DNA nanodevice transformable among three structural states
Abstract
In this work, we report the development and experimental validation of a coupled statistical thermodynamic model allowing prediction of the structural transitions executed by a novel DNA nanodevice, for quantitative operational design. The efficiency of target structure formation by this nanodevice, implemented with a bistable DNA molecule designed to transform between three distinct structures, is modeled by coupling the isolated equilibrium models for the individual structures. A peculiar behavior is predicted for this nanodevice, which forms the target structure within a limited temperature range by sensing thermal variations. The predicted thermal response is then validated via fluorescence measurements to quantitatively assess whether the nanodevice performs as designed. Agreement between predictions and experiment was substantial, with a 0.95 correlation for overall curve shape over a wide temperature range, from 30 C to 90 C. The obtained accuracy, which is comparable to that of conventional melting behavior prediction for DNA duplexes in isolation, ensures the applicability of the coupled model for illustrating general DNA reaction systems involving competitive duplex formation. Finally, tuning of the nanodevice using the current model towards design of a thermal band pass filter to control chemical circuits, as a novel function of DNA nanodevices is proposed.
Figures
 -
-  -
-  -
-  -
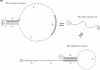
-  (5′–3′). The subsequence
(5′–3′). The subsequence  of length 13 nucleotides and its fully complementary subsequence
of length 13 nucleotides and its fully complementary subsequence  are indicated by shading.
are indicated by shading.  and
and  are poly-T subsequences of length 29 and 28 nt, respectively. Vertical lines represent base pairings. The fully melted coil form is depicted simply as a curved line. FAM and TAMRA fluorophores, attached for discrimination of the targeted hairpin structure, are indicated by F and T in gray and black circles, respectively. Upon formation of the targeted hairpin structure, the emission from FAM attached to the 5′ end of CH is preferentially quenched by the proximal TAMRA attached to the 3′ end.
are poly-T subsequences of length 29 and 28 nt, respectively. Vertical lines represent base pairings. The fully melted coil form is depicted simply as a curved line. FAM and TAMRA fluorophores, attached for discrimination of the targeted hairpin structure, are indicated by F and T in gray and black circles, respectively. Upon formation of the targeted hairpin structure, the emission from FAM attached to the 5′ end of CH is preferentially quenched by the proximal TAMRA attached to the 3′ end.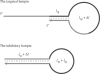
 was varied for simulation. The total length and length of the stem region of the targeted suboptimal hairpin structure,
was varied for simulation. The total length and length of the stem region of the targeted suboptimal hairpin structure,  were fixed to 95 nt and 12 bp, respectively (the same values as CH).
were fixed to 95 nt and 12 bp, respectively (the same values as CH).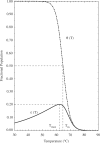
 reaches a maximum value of 0.20 at
reaches a maximum value of 0.20 at  = 62.3
= 62.3 C. For comparison, the dash-dotted curve indicates
C. For comparison, the dash-dotted curve indicates  , the predicted occupancy of the target hairpin structure when predictions are made via an isolated equilibrium model, along with the corresponding melting temperature,
, the predicted occupancy of the target hairpin structure when predictions are made via an isolated equilibrium model, along with the corresponding melting temperature,  . The characteristic temperatures of the two models are each indicated, with dashed lines added for clarity. The above prediction for
. The characteristic temperatures of the two models are each indicated, with dashed lines added for clarity. The above prediction for  , for a model system restricted to form the single target hairpin only, was experimentally validated in (28).
, for a model system restricted to form the single target hairpin only, was experimentally validated in (28).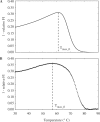
 ), by taking into account the unintended 14%-FRET quenching upon formation of the inhibitory hairpin. (B) The measured fluorescence footprint exhibited good agreement with the simulated fluorescence footprint, with a correlation of 0.95.
), by taking into account the unintended 14%-FRET quenching upon formation of the inhibitory hairpin. (B) The measured fluorescence footprint exhibited good agreement with the simulated fluorescence footprint, with a correlation of 0.95.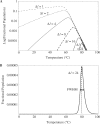
 are shown as semi-log plots. The width (FWHM) and maximum value obtained for FP decreased with increasing
are shown as semi-log plots. The width (FWHM) and maximum value obtained for FP decreased with increasing  . (B) Stringently limited formation of the targeted suboptimal hairpin was found after tuning. The predicted maximum value for FP and peak temperature were 0.0050% and 79.4°C, respectively, for
. (B) Stringently limited formation of the targeted suboptimal hairpin was found after tuning. The predicted maximum value for FP and peak temperature were 0.0050% and 79.4°C, respectively, for  24 bp.
24 bp.Similar articles
-
A DNA Nanodevice That Loads and Releases a Cargo with Hemoglobin-Like Allosteric Control and Cooperativity.Nano Lett. 2017 May 10;17(5):3225-3230. doi: 10.1021/acs.nanolett.7b00814. Epub 2017 Apr 11. Nano Lett. 2017. PMID: 28387120
-
Mapping the thermal behavior of DNA origami nanostructures.J Am Chem Soc. 2013 Apr 24;135(16):6165-76. doi: 10.1021/ja4000728. Epub 2013 Apr 12. J Am Chem Soc. 2013. PMID: 23537246
-
Cation-Activated Avidity for Rapid Reconfiguration of DNA Nanodevices.ACS Nano. 2018 Sep 25;12(9):9484-9494. doi: 10.1021/acsnano.8b04817. Epub 2018 Sep 4. ACS Nano. 2018. PMID: 30169013
-
Insights into the Structure and Energy of DNA Nanoassemblies.Molecules. 2020 Nov 24;25(23):5466. doi: 10.3390/molecules25235466. Molecules. 2020. PMID: 33255286 Free PMC article. Review.
-
Single-Molecule FRET: A Tool to Characterize DNA Nanostructures.Front Mol Biosci. 2022 Mar 7;9:835617. doi: 10.3389/fmolb.2022.835617. eCollection 2022. Front Mol Biosci. 2022. PMID: 35330798 Free PMC article. Review.
Cited by
-
The incipient denaturation mechanism of DNA.RSC Adv. 2022 Aug 17;12(36):23356-23365. doi: 10.1039/d2ra02480b. eCollection 2022 Aug 16. RSC Adv. 2022. PMID: 36090395 Free PMC article.
-
Structure and Dynamics of dsDNA in Cell-like Environments.Entropy (Basel). 2022 Nov 1;24(11):1587. doi: 10.3390/e24111587. Entropy (Basel). 2022. PMID: 36359677 Free PMC article. Review.
References
-
- Feldkamp U, Niemeyer CM. Rational design of DNA nanoarchitectures. Angew. Chem. Int. Ed. 2006;45:1856–1876. - PubMed
-
- Simmel FC. Three-dimensional nanoconstruction with DNA. Angew. Chem. Int. Ed. 2008;47:5884–5887. - PubMed
-
- Bath J, Turberfield AJ. DNA nanomachines. Nat. Nanotechnol. 2007;2:275–284. - PubMed
-
- Liedl T, Sobey TL, Simmel FC. DNA-based nanodevices. Nano Today. 2007;2:36–41.
-
- Yurke B, Turberfield AJ, Mills A.P., Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. - PubMed

