Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci
- PMID: 20385753
- PMCID: PMC2876548
- DOI: 10.1128/IAI.00688-09
Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci
Abstract
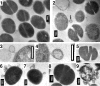
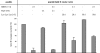
Human beta-defensin 3 (hBD3) is a highly charged (+11) cationic host defense peptide, produced by epithelial cells and neutrophils. hBD3 retains antimicrobial activity against a broad range of pathogens, including multiresistant Staphylococcus aureus, even under high-salt conditions. Whereas antimicrobial host defense peptides are assumed to act by permeabilizing cell membranes, the transcriptional response pattern of hBD3-treated staphylococcal cells resembled that of vancomycin-treated cells (V. Sass, U. Pag, A. Tossi, G. Bierbaum, and H. G. Sahl, Int. J. Med. Microbiol. 298:619-633, 2008) and suggested that inhibition of cell wall biosynthesis is a major component of the killing process. hBD3-treated cells, inspected by transmission electron microscopy, showed localized protrusions of cytoplasmic contents, and analysis of the intracellular pool of nucleotide-activated cell wall precursors demonstrated accumulation of the final soluble precursor, UDP-MurNAc-pentapeptide. Accumulation is typically induced by antibiotics that inhibit membrane-bound steps of cell wall biosynthesis and also demonstrates that hBD3 does not impair the biosynthetic capacity of cells and does not cause gross leakage of small cytoplasmic compounds. In in vitro assays of individual membrane-associated cell wall biosynthesis reactions (MraY, MurG, FemX, and penicillin-binding protein 2 [PBP2]), hBD3 inhibited those enzymes which use the bactoprenol-bound cell wall building block lipid II as a substrate; quantitative analysis suggested that hBD3 may stoichiometrically bind to lipid II. We report that binding of hBD3 to defined, lipid II-rich sites of cell wall biosynthesis may lead to perturbation of the biosynthesis machinery, resulting in localized lesions in the cell wall as demonstrated by electron microscopy. The lesions may then allow for osmotic rupture of cells when defensins are tested under low-salt conditions.
Figures

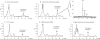


References
-
- Barker, H. C., N. Kinsella, A. Jaspe, T. Friedrich, and C. D. O'Connor. 2000. Formate protects stationary-phase Escherichia coli and Salmonella cells from killing by a cationic antimicrobial peptide. Mol. Microbiol. 35:1518-1529. - PubMed
-
- Bierbaum, G., and H. G. Sahl. 1989. Influence of cationic peptides on the activity of the autolytic endo-beta-N-acetylglucosaminidase of Staphylococcus simulans 22. FEMS Microbiol. Lett. 49:223-227. - PubMed
-
- Bowdish, D. M., D. J. Davidson, and R. E. Hancock. 2005. A re-evaluation of the role of host defence peptides in mammalian immunity. Curr. Protein Pept. Sci. 6:35-51. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

