Fetal liver hepatic progenitors are supportive stromal cells for hematopoietic stem cells
- PMID: 20385801
- PMCID: PMC2867886
- DOI: 10.1073/pnas.1003586107
Fetal liver hepatic progenitors are supportive stromal cells for hematopoietic stem cells
Abstract
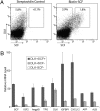
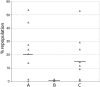
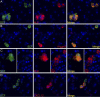
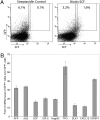
Previously we showed that the ~2% of fetal liver cells reactive with an anti-CD3epsilon monoclonal antibody support ex vivo expansion of both fetal liver and bone marrow hematopoietic stem cells (HSCs); these cells express two proteins important for HSC ex vivo expansion, IGF2, and angiopoietin-like 3. Here we show that these cells do not express any CD3 protein and are not T cells; rather, we purified these HSC-supportive stromal cells based on the surface phenotype of SCF(+)DLK(+). Competitive repopulating experiments show that SCF(+)DLK(+) cells support the maintenance of HSCs in ex vivo culture. These are the principal fetal liver cells that express not only angiopoietin-like 3 and IGF2, but also SCF and thrombopoietin, two other growth factors important for HSC expansion. They are also the principal fetal liver cells that express CXCL12, a factor required for HSC homing, and also alpha-fetoprotein (AFP), indicating that they are fetal hepatic stem or progenitor cells. Immunocytochemistry shows that >93% of the SCF(+) cells express DLK and Angptl3, and a portion of SCF(+) cells also expresses CXCL12. Thus SCF(+)DLK(+) cells are a highly homogenous population that express a complete set of factors for HSC expansion and are likely the primary stromal cells that support HSC expansion in the fetal liver.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Martinez-Agosto JA, Mikkola HK, Hartenstein V, Banerjee U. The hematopoietic stem cell and its niche: A comparative view. Genes Dev. 2007;21:3044–3060. - PubMed
-
- Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol. 2006;6:93–106. - PubMed
-
- Taichman RS. Blood and bone: Two tissues whose fates are intertwined to create the hematopoietic stem-cell niche. Blood. 2005;105:2631–2639. - PubMed
-
- Calvi LM, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. - PubMed
-
- Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

