Contrasts between adaptive coding and noncoding changes during human evolution
- PMID: 20385805
- PMCID: PMC2867918
- DOI: 10.1073/pnas.0911249107
Contrasts between adaptive coding and noncoding changes during human evolution
Abstract
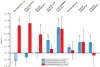
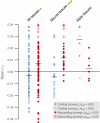
Changes in non-protein-coding regulatory DNA sequences have been proposed to play distinctive roles in adaptive evolution. We analyzed correlations between gene functions and evidence for positive selection in a common statistical framework across several large surveys of coding and noncoding sequences throughout the human genome. Strong correlations with both classifications in gene ontologies and measurements of gene expression indicate that neural development and function have adapted mainly through noncoding changes. In contrast, adaptation via coding changes is dominated by immunity, olfaction, and male reproduction. Genes with highly tissue-specific expression have undergone more adaptive coding changes, suggesting that pleiotropic constraints inhibit such changes in broadly expressed genes. In contrast, adaptive noncoding changes do not exhibit this pattern. Our findings underscore the probable importance of noncoding changes in the evolution of human traits, particularly cognitive traits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. - PubMed
-
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. - PubMed
-
- Stern DL. Evolutionary developmental biology and the problem of variation. Evolution. 2000;54:1079–1091. - PubMed
-
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

