Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration
- PMID: 20385819
- PMCID: PMC2867722
- DOI: 10.1073/pnas.0912702107
Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration
Abstract
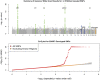
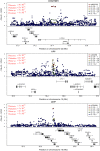
We executed a genome-wide association scan for age-related macular degeneration (AMD) in 2,157 cases and 1,150 controls. Our results validate AMD susceptibility loci near CFH (P < 10(-75)), ARMS2 (P < 10(-59)), C2/CFB (P < 10(-20)), C3 (P < 10(-9)), and CFI (P < 10(-6)). We compared our top findings with the Tufts/Massachusetts General Hospital genome-wide association study of advanced AMD (821 cases, 1,709 controls) and genotyped 30 promising markers in additional individuals (up to 7,749 cases and 4,625 controls). With these data, we identified a susceptibility locus near TIMP3 (overall P = 1.1 x 10(-11)), a metalloproteinase involved in degradation of the extracellular matrix and previously implicated in early-onset maculopathy. In addition, our data revealed strong association signals with alleles at two loci (LIPC, P = 1.3 x 10(-7); CETP, P = 7.4 x 10(-7)) that were previously associated with high-density lipoprotein cholesterol (HDL-c) levels in blood. Consistent with the hypothesis that HDL metabolism is associated with AMD pathogenesis, we also observed association with AMD of HDL-c-associated alleles near LPL (P = 3.0 x 10(-3)) and ABCA1 (P = 5.6 x 10(-4)). Multilocus analysis including all susceptibility loci showed that 329 of 331 individuals (99%) with the highest-risk genotypes were cases, and 85% of these had advanced AMD. Our studies extend the catalog of AMD associated loci, help identify individuals at high risk of disease, and provide clues about underlying cellular pathways that should eventually lead to new therapies.
Conflict of interest statement
Conflict of interest statement: A.O.E., J.J., M.A.P.-V., Y.P.C., A.K., W.K.S., A.A., E.A.P., M.M.D., D.E.W., J.M.S., J.L.H., M.B.G., G.R.A. and A.S. are listed as inventors on patents relating to the use of genetic data in age-related macular degeneration. S.G.S. has previously received research funding from Genentech and owns equity in Pfizer.
Figures

References
-
- Fisher SA, et al. Meta-analysis of genome scans of age-related macular degeneration. Hum Mol Genet. 2005;14:2257–2264. - PubMed
-
- Dewan A, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–992. - PubMed
-
- Rivera A, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–3236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 EY014801/EY/NEI NIH HHS/United States
- R01 EY009859/EY/NEI NIH HHS/United States
- R01 EY009769/EY/NEI NIH HHS/United States
- HG002651/HG/NHGRI NIH HHS/United States
- R01 EY012118/EY/NEI NIH HHS/United States
- HL084729/HL/NHLBI NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- EY016862/EY/NEI NIH HHS/United States
- R01 EY016862/EY/NEI NIH HHS/United States
- EY09859/EY/NEI NIH HHS/United States
- U10 EY012118/EY/NEI NIH HHS/United States
- EY-014458/EY/NEI NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
- R01 EY014458/EY/NEI NIH HHS/United States
- R01 EY014467/EY/NEI NIH HHS/United States
- U01 HL084729/HL/NHLBI NIH HHS/United States
- EY012118/EY/NEI NIH HHS/United States
- R01 HG002651/HG/NHGRI NIH HHS/United States
- Z99 EY999999/ImNIH/Intramural NIH HHS/United States
- EY007758/EY/NEI NIH HHS/United States
- R01 EY007758/EY/NEI NIH HHS/United States
- EY014467/EY/NEI NIH HHS/United States
- R01 EY011309/EY/NEI NIH HHS/United States
- P30-EY014801/EY/NEI NIH HHS/United States
- ZIA EY000489/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

