Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs
- PMID: 20386561
- PMCID: PMC3248744
- DOI: 10.1038/tpj.2010.28
Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs
Abstract
Cytochrome P450 3A4 (CYP3A4) metabolizes ∼50% of all clinically used drugs. Although CYP3A4 expression varies widely between individuals, the contribution of genetic factors remains uncertain. In this study, we measured allelic CYP3A4 heteronuclear RNA (hnRNA) and mRNA expression in 76 human liver samples heterozygous for at least one of eight marker SNPs and found marked allelic expression imbalance (1.6-6.3-fold) in 10/76 liver samples (13%). This was fully accounted for by an intron 6 SNP (rs35599367, C>T), which also affected mRNA expression in cell culture on minigene transfections. CYP3A4 mRNA level and enzyme activity in livers with CC genotype were 1.7- and 2.5-fold, respectively, greater than in CT and TT carriers. In 235 patients taking stable doses of atorvastatin, simvastatin, or lovastatin for lipid control, carriers of the T allele required significantly lower statin doses (0.2-0.6-fold, P=0.019) than non-T carriers for optimal lipid control. These results indicate that intron 6 SNP rs35599367 markedly affects expression of CYP3A4 and could serve as a biomarker for predicting response to CYP3A4-metabolized drugs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


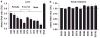



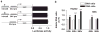
References
-
- Danielson PB. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3:561–597. - PubMed
-
- Westlind-Johnsson A, Malmebo S, Johansson A, Otter C, Andersson TB, Johansson I, et al. Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metab Dispos. 2003;31:755–761. - PubMed
-
- Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Inter-individual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270:414–423. - PubMed
-
- Westlind A, Lofberg L, Tindberg N, Andersson TB, Ingelman-Sundberg M. Interindividual differences in hepatic expression of CYP3A4: relationship to genetic polymorphism in the 5′-upstream regulatory region. Biochem Biophys Res Commun. 1999;259:201–205. - PubMed
-
- Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

