Structural characterization of CYP51 from Trypanosoma cruzi and Trypanosoma brucei bound to the antifungal drugs posaconazole and fluconazole
- PMID: 20386598
- PMCID: PMC2850312
- DOI: 10.1371/journal.pntd.0000651
Structural characterization of CYP51 from Trypanosoma cruzi and Trypanosoma brucei bound to the antifungal drugs posaconazole and fluconazole
Abstract
Background: Chagas Disease is the leading cause of heart failure in Latin America. Current drug therapy is limited by issues of both efficacy and severe side effects. Trypansoma cruzi, the protozoan agent of Chagas Disease, is closely related to two other major global pathogens, Leishmania spp., responsible for leishmaniasis, and Trypansoma brucei, the causative agent of African Sleeping Sickness. Both T. cruzi and Leishmania parasites have an essential requirement for ergosterol, and are thus vulnerable to inhibitors of sterol 14alpha-demethylase (CYP51), which catalyzes the conversion of lanosterol to ergosterol. Clinically employed anti-fungal azoles inhibit ergosterol biosynthesis in fungi, and specific azoles are also effective against both Trypanosoma and Leishmania parasites. However, modification of azoles to enhance efficacy and circumvent potential drug resistance has been problematic for both parasitic and fungal infections due to the lack of structural insights into drug binding.
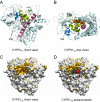
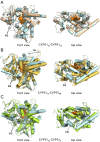
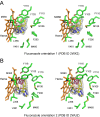
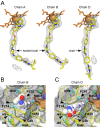
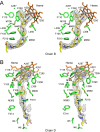
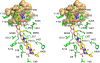
Methodology/principal findings: We have determined the crystal structures for CYP51 from T. cruzi (resolutions of 2.35 A and 2.27 A), and from the related pathogen T. brucei (resolutions of 2.7 A and 2.6 A), co-crystallized with the antifungal drugs fluconazole and posaconazole. Remarkably, both drugs adopt multiple conformations when binding the target. The fluconazole 2,4-difluorophenyl ring flips 180 degrees depending on the H-bonding interactions with the BC-loop. The terminus of the long functional tail group of posaconazole is bound loosely in the mouth of the hydrophobic substrate binding tunnel, suggesting that the major contribution of the tail to drug efficacy is for pharmacokinetics rather than in interactions with the target.
Conclusions/significance: The structures provide new insights into binding of azoles to CYP51 and mechanisms of potential drug resistance. Our studies define in structural detail the CYP51 therapeutic target in T. cruzi, and offer a starting point for rationally designed anti-Chagasic drugs with improved efficacy and reduced toxicity.
Conflict of interest statement
Matthew P. Jacobson is a consultant to Schrodinger, LLC.
Figures


Similar articles
-
Sterol 14alpha-demethylase (CYP51) as a therapeutic target for human trypanosomiasis and leishmaniasis.Curr Top Med Chem. 2011;11(16):2060-71. doi: 10.2174/156802611796575902. Curr Top Med Chem. 2011. PMID: 21619513 Free PMC article. Review.
-
Complexes of Trypanosoma cruzi sterol 14α-demethylase (CYP51) with two pyridine-based drug candidates for Chagas disease: structural basis for pathogen selectivity.J Biol Chem. 2013 Nov 1;288(44):31602-15. doi: 10.1074/jbc.M113.497990. Epub 2013 Sep 18. J Biol Chem. 2013. PMID: 24047900 Free PMC article.
-
Structural insights into inhibition of sterol 14alpha-demethylase in the human pathogen Trypanosoma cruzi.J Biol Chem. 2010 Aug 13;285(33):25582-90. doi: 10.1074/jbc.M110.133215. Epub 2010 Jun 8. J Biol Chem. 2010. PMID: 20530488 Free PMC article.
-
CYP51 from Trypanosoma cruzi: a phyla-specific residue in the B' helix defines substrate preferences of sterol 14alpha-demethylase.J Biol Chem. 2006 Feb 10;281(6):3577-85. doi: 10.1074/jbc.M510317200. Epub 2005 Nov 30. J Biol Chem. 2006. PMID: 16321980
-
CYP51 as drug targets for fungi and protozoan parasites: past, present and future.Parasitology. 2018 Dec;145(14):1820-1836. doi: 10.1017/S0031182018000562. Epub 2018 Apr 12. Parasitology. 2018. PMID: 29642960 Free PMC article. Review.
Cited by
-
Insights into Ergosterol Peroxide's Trypanocidal Activity.Biomolecules. 2019 Sep 12;9(9):484. doi: 10.3390/biom9090484. Biomolecules. 2019. PMID: 31547423 Free PMC article.
-
R-Configuration of 4-Aminopyridyl-Based Inhibitors of CYP51 Confers Superior Efficacy Against Trypanosoma cruzi.ACS Med Chem Lett. 2014 Jan 22;5(4):434-9. doi: 10.1021/ml500010m. eCollection 2014 Apr 10. ACS Med Chem Lett. 2014. PMID: 24900854 Free PMC article.
-
Rapid, Selection-Free, High-Efficiency Genome Editing in Protozoan Parasites Using CRISPR-Cas9 Ribonucleoproteins.mBio. 2017 Nov 7;8(6):e01788-17. doi: 10.1128/mBio.01788-17. mBio. 2017. PMID: 29114029 Free PMC article.
-
Recent Developments in Sterol 14-demethylase Inhibitors for Chagas Disease.Int J Parasitol Drugs Drug Resist. 2012 Dec;2:236-242. doi: 10.1016/j.ijpddr.2011.12.002. Int J Parasitol Drugs Drug Resist. 2012. PMID: 23277882 Free PMC article.
-
Progress and Prospects of Triazoles in Advanced Therapies for Parasitic Diseases.Trop Med Infect Dis. 2025 May 20;10(5):142. doi: 10.3390/tropicalmed10050142. Trop Med Infect Dis. 2025. PMID: 40423371 Free PMC article. Review.
References
-
- Chagas C. Nova trypanosomiase humana. Gaceta Medica da Bahia. 1909;40:433–440.
-
- World Health Organization. Geneva: WHO; 2002. Control of Chagas disease: second report of the WHO expert committee.
-
- Coura JR, De Castro SL. A Critical Review on Chagas Disease Chemotherapy. Memórias do Instituto Oswaldo Cruz. 2002;97:3–24. - PubMed
-
- Roberts CW, McLeod R, Rice DW, Ginger M, Chance ML, et al. Fatty acid and sterol metabolism: potential antimicrobial targets in apicomplexan and trypanosomatid parasitic protozoa. Mol Biochem Parasitol. 2003;126:129–142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

