Dynamics and control of diseases in networks with community structure
- PMID: 20386735
- PMCID: PMC2851561
- DOI: 10.1371/journal.pcbi.1000736
Dynamics and control of diseases in networks with community structure
Abstract
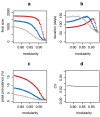
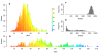
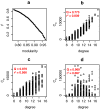
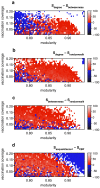
The dynamics of infectious diseases spread via direct person-to-person transmission (such as influenza, smallpox, HIV/AIDS, etc.) depends on the underlying host contact network. Human contact networks exhibit strong community structure. Understanding how such community structure affects epidemics may provide insights for preventing the spread of disease between communities by changing the structure of the contact network through pharmaceutical or non-pharmaceutical interventions. We use empirical and simulated networks to investigate the spread of disease in networks with community structure. We find that community structure has a major impact on disease dynamics, and we show that in networks with strong community structure, immunization interventions targeted at individuals bridging communities are more effective than those simply targeting highly connected individuals. Because the structure of relevant contact networks is generally not known, and vaccine supply is often limited, there is great need for efficient vaccination algorithms that do not require full knowledge of the network. We developed an algorithm that acts only on locally available network information and is able to quickly identify targets for successful immunization intervention. The algorithm generally outperforms existing algorithms when vaccine supply is limited, particularly in networks with strong community structure. Understanding the spread of infectious diseases and designing optimal control strategies is a major goal of public health. Social networks show marked patterns of community structure, and our results, based on empirical and simulated data, demonstrate that community structure strongly affects disease dynamics. These results have implications for the design of control strategies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
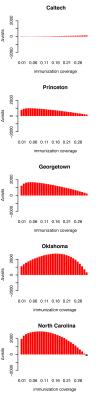
References
-
- Anderson RM, May RM. 1991. Infectious Diseases Of Humans, Dynamics and Control: Oxford Science Publications.
-
- Keeling MJ, Rohani P. 2008. Modeling Infectious Diseases in Humans and Animals: Princeton University Press.
-
- Diekmann O, Heesterbeek JAP. 2000. Mathematical Epidemiology of Infectious Diseases; Levin S, editor: John Wiley & Sons, Ltd.
-
- Anderson RM. Holmes K, editor. Transmission dynamics of sexually transmitted infections. 1999. pp. 25–37. Sexually Transmitted Diseases: McGraw-Hill.
-
- Morris M. Epidemiology and Social Networks - Modeling Structured Diffusion. Sociol Method Res. 1993;22:99–126.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

