Functional significance may underlie the taxonomic utility of single amino acid substitutions in conserved proteins
- PMID: 20386893
- PMCID: PMC2874023
- DOI: 10.1007/s00239-010-9338-y
Functional significance may underlie the taxonomic utility of single amino acid substitutions in conserved proteins
Abstract
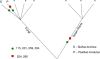
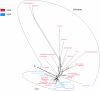
We hypothesized that some amino acid substitutions in conserved proteins that are strongly fixed by critical functional roles would show lineage-specific distributions. As an example of an archetypal conserved eukaryotic protein we considered the active site of beta-tubulin. Our analysis identified one amino acid substitution--beta-tubulin F224--which was highly lineage specific. Investigation of beta-tubulin for other phylogenetically restricted amino acids identified several with apparent specificity for well-defined phylogenetic groups. Intriguingly, none showed specificity for "supergroups" other than the unikonts. To understand why, we analysed the beta-tubulin Neighbor-Net and demonstrated a fundamental division between core beta-tubulins (plant-like) and divergent beta-tubulins (animal and fungal). F224 was almost completely restricted to the core beta-tubulins, while divergent beta-tubulins possessed Y224. Thus, our specific example offers insight into the restrictions associated with the co-evolution of beta-tubulin during the radiation of eukaryotes, underlining a fundamental dichotomy between F-type, core beta-tubulins and Y-type, divergent beta-tubulins. More broadly our study provides proof of principle for the taxonomic utility of critical amino acids in the active sites of conserved proteins.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

