Structural features promoting dioxygen production by Dechloromonas aromatica chlorite dismutase
- PMID: 20386942
- PMCID: PMC2909366
- DOI: 10.1007/s00775-010-0651-0
Structural features promoting dioxygen production by Dechloromonas aromatica chlorite dismutase
Abstract
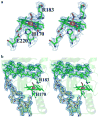
Chlorite dismutase (Cld) is a heme enzyme capable of rapidly and selectively decomposing chlorite (ClO(2) (-)) to Cl(-) and O(2). The ability of Cld to promote O(2) formation from ClO(2) (-) is unusual. Heme enzymes generally utilize ClO(2) (-) as an oxidant for reactions such as oxygen atom transfer to, or halogenation of, a second substrate. The X-ray crystal structure of Dechloromonas aromatica Cld co-crystallized with the substrate analogue nitrite (NO(2) (-)) was determined to investigate features responsible for this novel reactivity. The enzyme active site contains a single b-type heme coordinated by a proximal histidine residue. Structural analysis identified a glutamate residue hydrogen-bonded to the heme proximal histidine that may stabilize reactive heme species. A solvent-exposed arginine residue likely gates substrate entry to a tightly confined distal pocket. On the basis of the proposed mechanism of Cld, initial reaction of ClO(2) (-) within the distal pocket generates hypochlorite (ClO(-)) and a compound I intermediate. The sterically restrictive distal pocket probably facilitates the rapid rebound of ClO(-) with compound I forming the Cl(-) and O(2) products. Common to other heme enzymes, Cld is inactivated after a finite number of turnovers, potentially via the observed formation of an off-pathway tryptophanyl radical species through electron migration to compound I. Three tryptophan residues of Cld have been identified as candidates for this off-pathway radical. Finally, a juxtaposition of hydrophobic residues between the distal pocket and the enzyme surface suggests O(2) may have a preferential direction for exiting the active site.
Figures
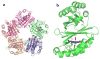






References
-
- Liu S, Suflita JM. Trends Biotechnol. 1993;11:344–352. - PubMed
-
- Wackett LP. J Biol Chem. 2004;279:41259–41262. - PubMed
-
- Wackett LP, Hershberger CD. Biocatalysis and biodegradation : microbial transformation of organic compounds. ASM Press; Washington: 2001.
-
- Oremland RS, Kulp TR, Blum JS, Hoeft SE, Baesman S, Miller LG, Stolz JF. Science. 2005;308:1305–1308. - PubMed
-
- Oremland RS, Stolz JF. Science. 2003;300:939–944. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

