Phloem small RNAs, nutrient stress responses, and systemic mobility
- PMID: 20388194
- PMCID: PMC2923538
- DOI: 10.1186/1471-2229-10-64
Phloem small RNAs, nutrient stress responses, and systemic mobility
Abstract
Background: Nutrient availabilities and needs have to be tightly coordinated between organs to ensure a balance between uptake and consumption for metabolism, growth, and defense reactions. Since plants often have to grow in environments with sub-optimal nutrient availability, a fine tuning is vital. To achieve this, information has to flow cell-to-cell and over long-distance via xylem and phloem. Recently, specific miRNAs emerged as a new type of regulating molecules during stress and nutrient deficiency responses, and miR399 was suggested to be a phloem-mobile long-distance signal involved in the phosphate starvation response.
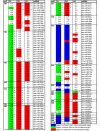
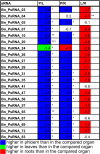
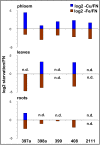
Results: We used miRNA microarrays containing all known plant miRNAs and a set of unknown small (s) RNAs earlier cloned from Brassica phloem sap 1, to comprehensively analyze the phloem response to nutrient deficiency by removing sulfate, copper or iron, respectively, from the growth medium. We show that phloem sap contains a specific set of sRNAs that is distinct from leaves and roots, and that the phloem also responds specifically to stress. Upon S and Cu deficiencies phloem sap reacts with an increase of the same miRNAs that were earlier characterized in other tissues, while no clear positive response to -Fe was observed. However, -Fe led to a reduction of Cu- and P-responsive miRNAs. We further demonstrate that under nutrient starvation miR399 and miR395 can be translocated through graft unions from wild type scions to rootstocks of the miRNA processing hen1-1 mutant. In contrast, miR171 was not transported. Translocation of miR395 led to a down-regulation of one of its targets in rootstocks, suggesting that this transport is of functional relevance, and that miR395, in addition to the well characterized miR399, could potentially act as a long-distance information transmitter.
Conclusions: Phloem sap contains a specific set of sRNAs, of which some specifically accumulate in response to nutrient deprivation. From the observation that miR395 and miR399 are phloem-mobile in grafting experiments we conclude that translocatable miRNAs might be candidates for information-transmitting molecules, but that grafting experiments alone are not sufficient to convincingly assign a signaling function.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

