Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps
- PMID: 20388197
- PMCID: PMC2884544
- DOI: 10.1186/gb-2010-11-4-r41
Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps
Abstract
Advances in sequencing technology allow genomes to be sequenced at vastly decreased costs. However, the assembled data frequently are highly fragmented with many gaps. We present a practical approach that uses Illumina sequences to improve draft genome assemblies by aligning sequences against contig ends and performing local assemblies to produce gap-spanning contigs. The continuity of a draft genome can thus be substantially improved, often without the need to generate new data.
Figures

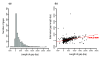
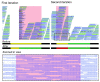

References
-
- Maccallum I, Przybylski D, Gnerre S, Burton J, Shlyakhter I, Gnirke A, Malek J, McKernan K, Ranade S, Shea TP, Williams L, Young S, Nusbaum C, Jaffe DB. ALLPATHS 2: small genomes assembled accurately and with high continuity from short paired reads. Genome Biol. 2009;10:R103. doi: 10.1186/gb-2009-10-10-r103. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

