Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry
- PMID: 20388209
- PMCID: PMC2861645
- DOI: 10.1186/1477-5956-8-22
Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry
Abstract
Background: Poly(ADP-ribose) polymerases (PARPs) catalyze the formation of poly(ADP-ribose) (pADPr), a post-translational modification involved in several important biological processes, namely surveillance of genome integrity, cell cycle progression, initiation of the DNA damage response, apoptosis, and regulation of transcription. Poly(ADP-ribose) glycohydrolase (PARG), on the other hand, catabolizes pADPr and thereby accounts for the transient nature of poly(ADP-ribosyl)ation. Our investigation of the interactomes of PARP-1, PARP-2, and PARG by affinity-purification mass spectrometry (AP-MS) aimed, on the one hand, to confirm current knowledge on these interactomes and, on the other hand, to discover new protein partners which could offer insights into PARPs and PARG functions.
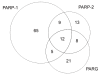
Results: PARP-1, PARP-2, and PARG were immunoprecipitated from human cells, and pulled-down proteins were separated by gel electrophoresis prior to in-gel trypsin digestion. Peptides were identified by tandem mass spectrometry. Our AP-MS experiments resulted in the identifications of 179 interactions, 139 of which are novel interactions. Gene Ontology analysis of the identified protein interactors points to five biological processes in which PARP-1, PARP-2 and PARG may be involved: RNA metabolism for PARP-1, PARP-2 and PARG; DNA repair and apoptosis for PARP-1 and PARP-2; and glycolysis and cell cycle for PARP-1.
Conclusions: This study reveals several novel protein partners for PARP-1, PARP-2 and PARG. It provides a global view of the interactomes of these proteins as well as a roadmap to establish the systems biology of poly(ADP-ribose) metabolism.
Figures



References
-
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. - DOI - PubMed
-
- Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

