Characterization of the mouse CP27 promoter and NF-Y mediated gene regulation
- PMID: 20388536
- PMCID: PMC2892892
- DOI: 10.1016/j.gene.2010.03.014
Characterization of the mouse CP27 promoter and NF-Y mediated gene regulation
Abstract
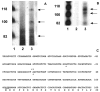
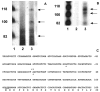
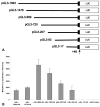
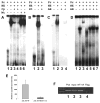
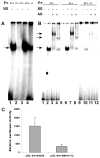
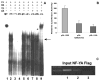
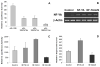
The cp27 gene is a highly conserved and unique gene with important roles related to craniofacial organogenesis. The present study is a first analysis of the CP27 promoter and its regulation. Here, we have cloned the promoter of the mouse cp27 gene, examined its transcriptional activity, and identified transcription factor binding sites in the proximal promoter region. Two major transcription start sites were mapped adjacent to exon 1. Promoter function analysis of the 5' flanking region by progressive 5' deletion mutations localized transcription repression elements between -1993bp and -969bp and several positive elements between -968bp and the preferred transcription start site. EMSA and functional studies indicated two function-cooperative CCAAT boxes and identified the NF-Y transcription factor as the CCAAT activator controlling transactivation of the CP27 promoter. In addition, this study demonstrated that for its effective binding and function, NF-Y required not only the minimal DNA segment length identified by deletion studies, but also a defined nucleotide sequence in the distal 3' flanking region of the CP27 proximal promoter CCAAT box. These results provide a basis for our understanding of the specific regulation of the cp27 gene in the NF-Y-mediated gene transcription network.
Copyright 2010 Elsevier B.V. All rights reserved.
Figures
Similar articles
-
NF-Y and USF1 transcription factor binding to CCAAT-box and E-box elements activates the CP27 promoter.Gene. 2011 Mar 1;473(2):92-9. doi: 10.1016/j.gene.2010.11.001. Epub 2010 Nov 13. Gene. 2011. PMID: 21078375 Free PMC article.
-
Transcriptional regulation of the mouse PNRC2 promoter by the nuclear factor Y (NFY) and E2F1.Gene. 2005 Nov 21;361:89-100. doi: 10.1016/j.gene.2005.07.012. Epub 2005 Sep 21. Gene. 2005. PMID: 16181749
-
Characterization of the human mitochondrial thiamine pyrophosphate transporter SLC25A19 minimal promoter: a role for NF-Y in regulating basal transcription.Gene. 2013 Oct 10;528(2):248-55. doi: 10.1016/j.gene.2013.06.073. Epub 2013 Jul 18. Gene. 2013. PMID: 23872534 Free PMC article.
-
Nuclear factor-Y (NF-Y) regulates transcription of mouse Dmrt7 gene by binding to tandem CCAAT boxes in its proximal promoter.Int J Biol Sci. 2010 Oct 25;6(7):655-64. doi: 10.7150/ijbs.6.655. Int J Biol Sci. 2010. PMID: 21060727 Free PMC article.
-
Characterization of the human EDF-1 minimal promoter: involvement of NFY and Sp1 in the regulation of basal transcription.Gene. 2006 Jun 7;374:87-95. doi: 10.1016/j.gene.2006.01.030. Epub 2006 Mar 29. Gene. 2006. PMID: 16567061
Cited by
-
The True Story of Yeti, the "Abominable" Heterochromatic Gene of Drosophila melanogaster.Front Physiol. 2019 Aug 22;10:1093. doi: 10.3389/fphys.2019.01093. eCollection 2019. Front Physiol. 2019. PMID: 31507454 Free PMC article. Review.
-
NF-Y and USF1 transcription factor binding to CCAAT-box and E-box elements activates the CP27 promoter.Gene. 2011 Mar 1;473(2):92-9. doi: 10.1016/j.gene.2010.11.001. Epub 2010 Nov 13. Gene. 2011. PMID: 21078375 Free PMC article.
References
-
- Bhattacharya A, Deng JM, Zhang Z, Behriger R, de Crombrugghe B, Maity SN. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. - PubMed
-
- Bi W, Wu L, Coustry F, de Crombrugghe B, Maity SN. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J Biol Chem. 1997;272:26562–26572. - PubMed
-
- Caretti G, Salsi V, Vecchi C, Imbriano C, Mantovani R. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J Biol Chem. 2003;278:30435–30440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

