The catalytic function of the Rev1 dCMP transferase is required in a lesion-specific manner for translesion synthesis and base damage-induced mutagenesis
- PMID: 20388628
- PMCID: PMC2926598
- DOI: 10.1093/nar/gkq225
The catalytic function of the Rev1 dCMP transferase is required in a lesion-specific manner for translesion synthesis and base damage-induced mutagenesis
Abstract
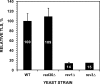
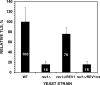
The Rev1-Polzeta pathway is believed to be the major mechanism of translesion DNA synthesis and base damage-induced mutagenesis in eukaryotes. While it is widely believed that Rev1 plays a non-catalytic function in translesion synthesis, the role of its dCMP transferase activity remains uncertain. To determine the relevance of its catalytic function in translesion synthesis, we separated the Rev1 dCMP transferase activity from its non-catalytic function in yeast. This was achieved by mutating two conserved amino acid residues in the catalytic domain of Rev1, i.e. D467A/E468A, where its catalytic function was abolished but its non-catalytic function remained intact. In this mutant strain, whereas translesion synthesis and mutagenesis of UV radiation were fully functional, those of a site-specific 1,N(6)-ethenoadenine were severely deficient. Specifically, the predominant A-->G mutations resulting from C insertion opposite the lesion were abolished. Therefore, translesion synthesis and mutagenesis of 1,N(6)-ethenoadenine require the catalytic function of the Rev1 dCMP transferase, in contrast to those of UV lesions, which only require the non-catalytic function of Rev1. These results show that the catalytic function of the Rev1 dCMP transferase is required in a lesion-specific manner for translesion synthesis and base damage-induced mutagenesis.
Figures




Similar articles
-
Translesion synthesis of acetylaminofluorene-dG adducts by DNA polymerase zeta is stimulated by yeast Rev1 protein.Nucleic Acids Res. 2004 Feb 11;32(3):1122-30. doi: 10.1093/nar/gkh279. Print 2004. Nucleic Acids Res. 2004. PMID: 14960722 Free PMC article.
-
A non-catalytic function of Rev1 in translesion DNA synthesis and mutagenesis is mediated by its stable interaction with Rad5.DNA Repair (Amst). 2013 Jan 1;12(1):27-37. doi: 10.1016/j.dnarep.2012.10.003. Epub 2012 Nov 9. DNA Repair (Amst). 2013. PMID: 23142547
-
Poleta, Polzeta and Rev1 together are required for G to T transversion mutations induced by the (+)- and (-)-trans-anti-BPDE-N2-dG DNA adducts in yeast cells.Nucleic Acids Res. 2006 Jan 13;34(2):417-25. doi: 10.1093/nar/gkj446. Print 2006. Nucleic Acids Res. 2006. PMID: 16415180 Free PMC article.
-
Translesion synthesis by the UmuC family of DNA polymerases.Mutat Res. 2001 Jul 12;486(2):59-70. doi: 10.1016/s0921-8777(01)00089-1. Mutat Res. 2001. PMID: 11425512 Review.
-
Roles of mutagenic translesion synthesis in mammalian genome stability, health and disease.DNA Repair (Amst). 2015 May;29:56-64. doi: 10.1016/j.dnarep.2015.01.001. Epub 2015 Jan 21. DNA Repair (Amst). 2015. PMID: 25655219 Review.
Cited by
-
The DNA polymerase activity of Saccharomyces cerevisiae Rev1 is biologically significant.Genetics. 2011 Jan;187(1):21-35. doi: 10.1534/genetics.110.124172. Epub 2010 Oct 26. Genetics. 2011. PMID: 20980236 Free PMC article.
-
Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.Nat Commun. 2017 Oct 17;8(1):965. doi: 10.1038/s41467-017-01013-5. Nat Commun. 2017. PMID: 29042535 Free PMC article.
-
Sml1 Inhibits the DNA Repair Activity of Rev1 in Saccharomyces cerevisiae during Oxidative Stress.Appl Environ Microbiol. 2020 Mar 18;86(7):e02838-19. doi: 10.1128/AEM.02838-19. Print 2020 Mar 18. Appl Environ Microbiol. 2020. PMID: 32005731 Free PMC article.
-
Translesion synthesis polymerases are dispensable for C. elegans reproduction but suppress genome scarring by polymerase theta-mediated end joining.PLoS Genet. 2020 Apr 24;16(4):e1008759. doi: 10.1371/journal.pgen.1008759. eCollection 2020 Apr. PLoS Genet. 2020. PMID: 32330130 Free PMC article.
-
Genomic and functional integrity of the hematopoietic system requires tolerance of oxidative DNA lesions.Blood. 2017 Sep 28;130(13):1523-1534. doi: 10.1182/blood-2017-01-764274. Epub 2017 Aug 21. Blood. 2017. PMID: 28827409 Free PMC article.
References
-
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. - PubMed
-
- Wang Z. Mechanism of bypass polymerases in eukaryotes. In: Siede W, Kow YW, Doetsch PW, editors. DNA Damage Recognition. New York, NY: Taylor & Francis Group; 2006. pp. 507–528.
-
- Xie Z, Zhang Y, Guliaev AB, Shen H, Hang B, Singer B, Wang Z. The p-benzoquinone DNA adducts derived from benzene are highly mutagenic. DNA Repair. 2005;4:1399–1409. - PubMed
-
- Xie Z, Braithwaite E, Guo D, Zhao B, Geacintov NE, Wang Z. Mutagenesis of benzo[a]pyrene diol epoxide in yeast: requirement for DNA polymerase ζ and involvement of DNA polymerase η. Biochemistry. 2003;42:11253–11262. - PubMed
-
- Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol. 2000;37:549–554. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

