The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction
- PMID: 20388712
- PMCID: PMC2881810
- DOI: 10.1074/jbc.M110.105668
The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction
Abstract
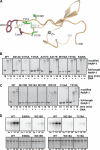
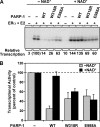
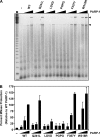
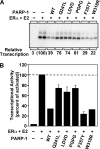
PARP-1 is involved in multiple cellular processes, including transcription, DNA repair, and apoptosis. PARP-1 attaches ADP-ribose units to target proteins, including itself as a post-translational modification that can change the biochemical properties of target proteins and mediate recruitment of proteins to sites of poly(ADP-ribose) synthesis. Independent of its catalytic activity, PARP-1 binds to chromatin and promotes compaction affecting RNA polymerase II transcription. PARP-1 has a modular structure composed of six independent domains. Two homologous zinc fingers, Zn1 and Zn2, form the DNA-binding module. Zn1-Zn2 binding to DNA breaks triggers catalytic activity. Recently, we have identified a third zinc binding domain in PARP-1, the Zn3 domain, which is essential for DNA-dependent PARP-1 activity. The crystal structure of the Zn3 domain revealed a novel zinc-ribbon fold and a homodimeric Zn3 structure that formed in the crystal lattice. Structure-guided mutagenesis was used here to investigate the roles of these two features of the Zn3 domain. Our results indicate that the zinc-ribbon fold of the Zn3 domain mediates an interdomain contact crucial to assembly of the DNA-activated conformation of PARP-1. In contrast, residues located at the Zn3 dimer interface are not required for DNA-dependent activation but rather make important contributions to the chromatin compaction activity of PARP-1. Thus, the Zn3 domain has dual roles in regulating the functions of PARP-1.
Figures
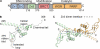



Similar articles
-
Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity.J Biol Chem. 2011 Mar 4;286(9):7149-60. doi: 10.1074/jbc.M110.175190. Epub 2010 Dec 23. J Biol Chem. 2011. PMID: 21183686 Free PMC article.
-
Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity.J Biol Chem. 2011 Mar 25;286(12):10690-701. doi: 10.1074/jbc.M110.202507. Epub 2011 Jan 13. J Biol Chem. 2011. PMID: 21233213 Free PMC article.
-
A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation.J Biol Chem. 2008 Feb 15;283(7):4105-14. doi: 10.1074/jbc.M708558200. Epub 2007 Nov 30. J Biol Chem. 2008. PMID: 18055453
-
PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis.Curr Opin Struct Biol. 2013 Feb;23(1):134-43. doi: 10.1016/j.sbi.2013.01.003. Epub 2013 Jan 16. Curr Opin Struct Biol. 2013. PMID: 23333033 Free PMC article. Review.
-
Regulation of chromatin structure and chromatin-dependent transcription by poly(ADP-ribose) polymerase-1: possible targets for drug-based therapies.Subcell Biochem. 2007;41:45-69. doi: 10.1007/1-4020-5466-1_3. Subcell Biochem. 2007. PMID: 17484123 Review.
Cited by
-
Structural basis for lack of ADP-ribosyltransferase activity in poly(ADP-ribose) polymerase-13/zinc finger antiviral protein.J Biol Chem. 2015 Mar 20;290(12):7336-44. doi: 10.1074/jbc.M114.630160. Epub 2015 Jan 29. J Biol Chem. 2015. PMID: 25635049 Free PMC article.
-
Functional roles of ADP-ribosylation writers, readers and erasers.Front Cell Dev Biol. 2022 Aug 11;10:941356. doi: 10.3389/fcell.2022.941356. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36035988 Free PMC article. Review.
-
Crystal structure of human ADP-ribose transferase ARTD15/PARP16 reveals a novel putative regulatory domain.J Biol Chem. 2012 Jul 13;287(29):24077-81. doi: 10.1074/jbc.M112.379289. Epub 2012 Jun 1. J Biol Chem. 2012. PMID: 22661712 Free PMC article.
-
Poly(ADP-ribose) polymerase-1 antagonizes DNA resection at double-strand breaks.Nat Commun. 2019 Jul 4;10(1):2954. doi: 10.1038/s41467-019-10741-9. Nat Commun. 2019. PMID: 31273204 Free PMC article.
-
The dynamic process of covalent and non-covalent PARylation in the maintenance of genome integrity: a focus on PARP inhibitors.NAR Cancer. 2023 Aug 21;5(3):zcad043. doi: 10.1093/narcan/zcad043. eCollection 2023 Sep. NAR Cancer. 2023. PMID: 37609662 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

