Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation
- PMID: 20388916
- PMCID: PMC2982149
- DOI: 10.1126/scisignal.2000594
Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation
Erratum in
- Sci Signal. 2010;3(123):er6
Abstract
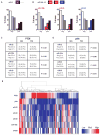
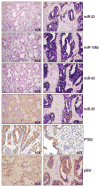
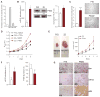
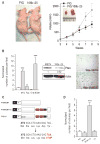
PTEN (phosphatase and tensin homolog deleted on chromosome 10) is a tumor suppressor that antagonizes signaling through the phosphatidylinositol 3-kinase-Akt pathway. We have demonstrated that subtle decreases in PTEN abundance can have critical consequences for tumorigenesis. Here, we used a computational approach to identify miR-22, miR-25, and miR-302 as three PTEN-targeting microRNA (miRNA) families found within nine genomic loci. We showed that miR-22 and the miR-106b~25 cluster are aberrantly overexpressed in human prostate cancer, correlate with abundance of the miRNA processing enzyme DICER, and potentiate cellular transformation both in vitro and in vivo. We demonstrated that the intronic miR-106b~25 cluster cooperates with its host gene MCM7 in cellular transformation both in vitro and in vivo, so that the concomitant overexpression of MCM7 and the miRNA cluster triggers prostatic intraepithelial neoplasia in transgenic mice. Therefore, the MCM7 gene locus delivers two simultaneous oncogenic insults when amplified or overexpressed in human cancer. Thus, we have uncovered a proto-oncogenic miRNA-dependent network for PTEN regulation and defined the MCM7 locus as a critical factor in initiating prostate tumorigenesis.
Conflict of interest statement
Figures






References
-
-
Nomenclature: PTEN, human protein; PTEN, human mRNA and gene; Pten, mouse protein; Pten, mouse mRNA and gene; DICER, human protein; DICER, human mRNA or gene; MCM7, human protein; MCM7, human mRNA and gene; miR-X, mature miRNA; miR-X, miRNA gene.
-
-
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. - PubMed
-
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

