Molecular identification of NAT8 as the enzyme that acetylates cysteine S-conjugates to mercapturic acids
- PMID: 20392701
- PMCID: PMC2881811
- DOI: 10.1074/jbc.M110.110924
Molecular identification of NAT8 as the enzyme that acetylates cysteine S-conjugates to mercapturic acids
Abstract
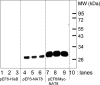
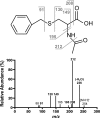
Our goal was to identify the reaction catalyzed by NAT8 (N-acetyltransferase 8), a putative N-acetyltransferase homologous to the enzyme (NAT8L) that produces N-acetylaspartate in brain. The almost exclusive expression of NAT8 in kidney and liver and its predicted association with the endoplasmic reticulum suggested that it was cysteinyl-S-conjugate N-acetyltransferase, the microsomal enzyme that catalyzes the last step of mercapturic acid formation. In agreement, HEK293T extracts of cells overexpressing NAT8 catalyzed the N-acetylation of S-benzyl-L-cysteine and leukotriene E(4), two cysteine conjugates, but were inactive on other physiological amines or amino acids. Confocal microscopy indicated that NAT8 was associated with the endoplasmic reticulum. Neither of the two frequent single nucleotide polymorphisms found in NAT8, E104K nor F143S, changed the enzymatic activity or the expression of the protein by >or=2-fold, whereas a mutation (R149K) replacing an extremely conserved arginine suppressed the activity. Sequencing of genomic DNA and EST clones corresponding to the NAT8B gene, which resulted from duplication of the NAT8 gene in the primate lineage, disclosed the systematic presence of a premature stop codon at codon 16. Furthermore, truncated NAT8B and NAT8 proteins starting from the following methionine (Met-25) showed no cysteinyl-S-conjugate N-acetyltransferase activity when transfected in HEK293T cells. Taken together, these findings indicate that NAT8 is involved in mercapturic acid formation and confirm that NAT8B is an inactive gene in humans. NAT8 homologues are found in all vertebrate genomes, where they are often encoded by multiple, tandemly repeated genes as many other genes encoding xenobiotic metabolism enzymes.
Figures





References
-
- Wiame E., Tyteca D., Pierrot N., Collard F., Amyere M., Noel G., Desmedt J., Nassogne M. C., Vikkula M., Octave J. N., Vincent M. F., Courtoy P. J., Boltshauser E., van Schaftingen E. (2010) Biochem. J. 425, 127–136 - PubMed
-
- Ozaki K., Fujiwara T., Nakamura Y., Takahashi E. (1998) J. Hum. Genet. 43, 255–258 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

