A novel F-box protein is required for caspase activation during cellular remodeling in Drosophila
- PMID: 20392747
- PMCID: PMC2860250
- DOI: 10.1242/dev.050088
A novel F-box protein is required for caspase activation during cellular remodeling in Drosophila
Abstract
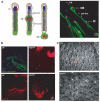
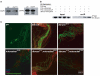
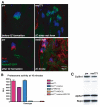
Terminal differentiation of male germ cells in Drosophila and mammals requires extensive cytoarchitectural remodeling, the elimination of many organelles, and a large reduction in cell volume. The associated process, termed spermatid individualization, is facilitated by the apoptotic machinery, including caspases, but does not result in cell death. From a screen for genes defective in caspase activation in this system, we isolated a novel F-box protein, which we termed Nutcracker, that is strictly required for caspase activation and sperm differentiation. Nutcracker interacts through its F-box domain with members of a Cullin-1-based ubiquitin ligase complex (SCF): Cullin-1 and SkpA. This ubiquitin ligase does not regulate the stability of the caspase inhibitors DIAP1 and DIAP2, but physically binds Bruce, a BIR-containing giant protein involved in apoptosis regulation. Furthermore, nutcracker mutants disrupt proteasome activity without affecting their distribution. These findings define a new SCF complex required for caspase activation during sperm differentiation and highlight the role of regulated proteolysis during this process.
Figures




Similar articles
-
A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila.PLoS Biol. 2007 Oct;5(10):e251. doi: 10.1371/journal.pbio.0050251. PLoS Biol. 2007. PMID: 17880263 Free PMC article.
-
A conserved F box regulatory complex controls proteasome activity in Drosophila.Cell. 2011 Apr 29;145(3):371-82. doi: 10.1016/j.cell.2011.03.021. Cell. 2011. PMID: 21529711 Free PMC article.
-
Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization.PLoS Biol. 2004 Jan;2(1):E15. doi: 10.1371/journal.pbio.0020015. Epub 2003 Dec 15. PLoS Biol. 2004. PMID: 14737191 Free PMC article.
-
Drosophila morgue and the intersection between protein ubiquitination and programmed cell death.Apoptosis. 2003 Mar;8(2):129-39. doi: 10.1023/a:1022914524601. Apoptosis. 2003. PMID: 12766473 Review.
-
Nerve-racking - apoptotic and non-apoptotic roles of caspases in the nervous system of Drosophila.Eur J Neurosci. 2016 Jul;44(1):1683-90. doi: 10.1111/ejn.13213. Epub 2016 Mar 19. Eur J Neurosci. 2016. PMID: 26900934 Review.
Cited by
-
The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy.Nat Neurosci. 2013 Sep;16(9):1257-65. doi: 10.1038/nn.3489. Epub 2013 Aug 11. Nat Neurosci. 2013. PMID: 23933751 Free PMC article.
-
Stallion sperm transcriptome comprises functionally coherent coding and regulatory RNAs as revealed by microarray analysis and RNA-seq.PLoS One. 2013;8(2):e56535. doi: 10.1371/journal.pone.0056535. Epub 2013 Feb 11. PLoS One. 2013. PMID: 23409192 Free PMC article.
-
Mulet (mlt) encodes a tubulin-binding cofactor E-like homolog required for spermatid individualization in Drosophila melanogaster.Fly (Austin). 2012 Oct-Dec;6(4):261-72. doi: 10.4161/fly.21533. Epub 2012 Aug 13. Fly (Austin). 2012. PMID: 22885996 Free PMC article.
-
Role of Selective Autophagy in Spermatogenesis and Male Fertility.Cells. 2020 Nov 23;9(11):2523. doi: 10.3390/cells9112523. Cells. 2020. PMID: 33238415 Free PMC article. Review.
-
Separating from the pack: Molecular mechanisms of Drosophila spermatid individualization.Spermatogenesis. 2015 May 21;5(2):e1041345. doi: 10.1080/21565562.2015.1041345. eCollection 2015 May-Aug. Spermatogenesis. 2015. PMID: 26413413 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

