Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae
- PMID: 20392820
- PMCID: PMC2926614
- DOI: 10.1093/nar/gkq260
Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae
Abstract
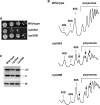
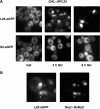
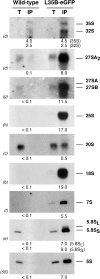
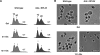
Ribosome synthesis involves the concomitance of pre-rRNA processing and ribosomal protein assembly. In eukaryotes, this is a complex process that requires the participation of specific sequences and structures within the pre-rRNAs, at least 200 trans-acting factors and the ribosomal proteins. There is little information on the function of individual 60S ribosomal proteins in ribosome synthesis. Herein, we have analysed the contribution of ribosomal protein L35 in ribosome biogenesis. In vivo depletion of L35 results in a deficit in 60S ribosomal subunits and the appearance of half-mer polysomes. Pulse-chase, northern hybridization and primer extension analyses show that processing of the 27SB to 7S pre-rRNAs is strongly delayed upon L35 depletion. Most likely as a consequence of this, release of pre-60S ribosomal particles from the nucleolus to the nucleoplasm is also blocked. Deletion of RPL35A leads to similar although less pronounced phenotypes. Moreover, we show that L35 assembles in the nucleolus and binds to early pre-60S ribosomal particles. Finally, flow cytometry analysis indicated that L35-depleted cells mildly delay the G1 phase of the cell cycle. We conclude that L35 assembly is a prerequisite for the efficient cleavage of the internal transcribed spacer 2 at site C(2).
Figures




References
-
- Steitz TA. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell. Biol. 2008;9:242–253. - PubMed
-
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. - PubMed
-
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

