Quantitative proteomics analysis reveals BAG3 as a potential target to suppress severe acute respiratory syndrome coronavirus replication
- PMID: 20392858
- PMCID: PMC2876644
- DOI: 10.1128/JVI.00213-10
Quantitative proteomics analysis reveals BAG3 as a potential target to suppress severe acute respiratory syndrome coronavirus replication
Abstract
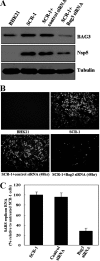
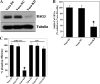
The discovery of a novel coronavirus (CoV) as the causative agent of severe acute respiratory syndrome (SARS) has highlighted the need for a better understanding of CoV replication. The replication of SARS-CoV is highly dependent on host cell factors. However, relatively little is known about the cellular proteome changes that occur during SARS-CoV replication. Recently, we developed a cell line expressing a SARS-CoV subgenomic replicon and used it to screen inhibitors of SARS-CoV replication. To identify host proteins important for SARS-CoV RNA replication, the protein profiles of the SARS-CoV replicon cells and parental BHK21 cells were compared using a quantitative proteomic strategy termed "stable-isotope labeling by amino acids in cell culture-mass spectrometry" (SILAC-MS). Our results revealed that, among the 1,081 host proteins quantified in both forward and reverse SILAC measurements, 74 had significantly altered levels of expression. Of these, significantly upregulated BCL2-associated athanogene 3 (BAG3) was selected for further functional studies. BAG3 is involved in a wide variety of cellular processes, including cell survival, cellular stress response, proliferation, migration, and apoptosis. Our results show that inhibition of BAG3 expression by RNA interference led to significant suppression of SARS-CoV replication, suggesting the possibility that upregulation of BAG3 may be part of the machinery that SARS-CoV relies on for replication. By correlating the proteomic data with these functional studies, the findings of this study provide important information for understanding SARS-CoV replication.
Figures


Similar articles
-
Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2'-o-methyltransferase activity.J Virol. 2014 Apr;88(8):4251-64. doi: 10.1128/JVI.03571-13. Epub 2014 Jan 29. J Virol. 2014. PMID: 24478444 Free PMC article.
-
High-throughput assay using a GFP-expressing replicon for SARS-CoV drug discovery.Antiviral Res. 2008 Nov;80(2):107-13. doi: 10.1016/j.antiviral.2008.05.005. Epub 2008 Jun 13. Antiviral Res. 2008. PMID: 18584889 Free PMC article.
-
Severe Acute Respiratory Syndrome Coronavirus ORF7a Inhibits Bone Marrow Stromal Antigen 2 Virion Tethering through a Novel Mechanism of Glycosylation Interference.J Virol. 2015 Dec;89(23):11820-33. doi: 10.1128/JVI.02274-15. Epub 2015 Sep 16. J Virol. 2015. PMID: 26378163 Free PMC article.
-
Signal transduction in SARS-CoV-infected cells.Ann N Y Acad Sci. 2007 Apr;1102(1):86-95. doi: 10.1196/annals.1408.006. Ann N Y Acad Sci. 2007. PMID: 17470913 Free PMC article. Review.
-
Unique SARS-CoV protein nsp1: bioinformatics, biochemistry and potential effects on virulence.Trends Microbiol. 2007 Feb;15(2):51-3. doi: 10.1016/j.tim.2006.12.005. Epub 2007 Jan 4. Trends Microbiol. 2007. PMID: 17207625 Free PMC article. Review.
Cited by
-
Nucleolin mediates SARS-CoV-2 replication and viral-induced apoptosis of host cells.Antiviral Res. 2023 Mar;211:105550. doi: 10.1016/j.antiviral.2023.105550. Epub 2023 Feb 3. Antiviral Res. 2023. PMID: 36740097 Free PMC article.
-
Identification of host factors involved in coronavirus replication by quantitative proteomics analysis.Proteomics. 2011 Jan;11(1):64-80. doi: 10.1002/pmic.201000309. Epub 2010 Dec 6. Proteomics. 2011. PMID: 21182195 Free PMC article.
-
Bcl2-associated athanogene 3 interactome analysis reveals a new role in modulating proteasome activity.Mol Cell Proteomics. 2013 Oct;12(10):2804-19. doi: 10.1074/mcp.M112.025882. Epub 2013 Jul 3. Mol Cell Proteomics. 2013. PMID: 23824909 Free PMC article.
-
Mitochondrial ribosomal stress in lung diseases.Am J Physiol Lung Cell Mol Physiol. 2022 Apr 1;322(4):L507-L517. doi: 10.1152/ajplung.00078.2021. Epub 2021 Dec 7. Am J Physiol Lung Cell Mol Physiol. 2022. PMID: 34873929 Free PMC article. Review.
-
Viral proteomics: the emerging cutting-edge of virus research.Sci China Life Sci. 2011 Jun;54(6):502-12. doi: 10.1007/s11427-011-4177-7. Epub 2011 Jun 26. Sci China Life Sci. 2011. PMID: 21706410 Free PMC article. Review.
References
-
- Aitken, A. 2006. 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 16:162-172. - PubMed
-
- Blagoev, B., S. E. Ong, I. Kratchmarova, and M. Mann. 2004. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 22:1139-1145. - PubMed
-
- Bonelli, P., A. Petrella, A. Rosati, M. F. Romano, R. Lerose, M. G. Pagliuca, T. Amelio, M. Festa, G. Martire, S. Venuta, M. C. Turco, and A. Leone. 2004. BAG3 protein regulates stress-induced apoptosis in normal and neoplastic leukocytes. Leukemia 18:358-360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

