Development and evaluation of an ompA quantitative real-time PCR assay for Chlamydia trachomatis serovar determination
- PMID: 20392903
- PMCID: PMC2884500
- DOI: 10.1128/JCM.02308-09
Development and evaluation of an ompA quantitative real-time PCR assay for Chlamydia trachomatis serovar determination
Abstract
Knowledge of circulating Chlamydia trachomatis serovars can be beneficial for sexual network surveillance, monitoring treatment success, and associating specific clinical manifestations. Typically, C. trachomatis serovars are predicted by nucleotide sequencing of four variable domains within the ompA gene. However, sequencing procedures can be labor-intensive, are not readily available, and can lack the capacity to identify multiple serovars. This study describes the development and evaluation of a quantitative real-time PCR (qPCR) test algorithm for the rapid prediction of C. trachomatis serovars, including ocular (A to C) and anogenital (D to L3) strains. This test comprises a primary qPCR to confirm C. trachomatis positivity and the phylogenetic group(s) present and a secondary set of qPCRs to determine specific serovars. Cell culture isolates from 15 prototypic C. trachomatis serovars were correctly identified using this assay, with no cross-reactivity observed among serovars or with other common pathogenic microorganisms. Five hundred clinical specimens (previously diagnosed as being C. trachomatis positive) were evaluated by qPCR, with their results compared to results obtained by conventional sequencing. The qPCR identified 88.9% (423/476) complete matches (95% confidence interval [CI], 86 to 92%) of serovars compared to the results obtained using the sequence-based approach. Among the anogenital specimens, 2.4% (12/494) (95% CI, 1.3 to 4.2%) contained multiple serovars, categorized as single-serovar infections by conventional sequencing. Overall, this test exhibited high discriminatory success for predicting C. trachomatis serovars, particularly among anogenital infections. This is the first report of a qPCR typing assay offering differentiation of C. trachomatis serovars associated with both anogenital and ocular diseases.
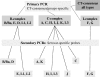
Figures

References
-
- Bandea, C. I., K. Kubota, T. M. Brown, P. H. Kilmarx, V. Bhullar, S. Yanpaisarn, P. Chaisilwattana, W. Siriwasin, and C. M. Black. 2001. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex. Transm. Infect. 77:419-422. - PMC - PubMed
-
- Cates, W., Jr., and J. N. Wasserheit. 1991. Genital chlamydial infections: epidemiology and reproductive sequelae. Am. J. Obstet. Gynecol. 164:1771-1781. - PubMed
-
- Garland, S. M., and B. Johnson. 1989. Chlamydia trachomatis infections—The Royal Women's Hospital experience. Med. J. Aust. 150:174-177. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

