Molecular identification of veterinary yeast isolates by use of sequence-based analysis of the D1/D2 region of the large ribosomal subunit
- PMID: 20392917
- PMCID: PMC2884494
- DOI: 10.1128/JCM.02306-09
Molecular identification of veterinary yeast isolates by use of sequence-based analysis of the D1/D2 region of the large ribosomal subunit
Abstract
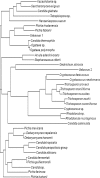
Conventional methods of yeast identification are often time-consuming and difficult; however, recent studies of sequence-based identification methods have shown promise. Additionally, little is known about the diversity of yeasts identified from various animal species in veterinary diagnostic laboratories. Therefore, in this study, we examined three methods of identification by using 109 yeast samples isolated during a 1-year period from veterinary clinical samples. Comparison of the three methods-traditional substrate assimilation, fatty acid profile analysis, and sequence-based analysis of the region spanning the D1 and D2 regions (D1/D2) of the large ribosomal subunit-showed that sequence analysis provided the highest percent identification among the three. Sequence analysis identified 87% of isolates to the species level, whereas substrate assimilation and fatty acid profile analysis identified only 54% and 47%, respectively. Less-stringent criteria for identification increased the percentage of isolates identified to 98% for sequence analysis, 62% for substrate assimilation, and 55% for fatty acid profile analysis. We also found that sequence analysis of the internal transcribed spacer 2 (ITS2) region provided further identification for 36% of yeast not identified to the species level by D1/D2 sequence analysis. Additionally, we identified a large variety of yeast from animal sources, with at least 30 different species among the isolates tested, and with the majority not belonging to the common Candida spp., such as C. albicans, C. glabrata, C. tropicalis, and the C. parapsilosis group. Thus, we determined that sequence analysis of the D1/D2 region was the best method for identification of the variety of yeasts found in a veterinary population.
Figures

Similar articles
-
Identification and antifungal susceptibility of a large collection of yeast strains isolated in Tunisian hospitals.Med Mycol. 2013 Oct;51(7):737-46. doi: 10.3109/13693786.2013.800239. Epub 2013 Jun 14. Med Mycol. 2013. PMID: 23768242
-
[Rapid identification of clinical yeast species by single-strand conformation polymorphism analysis of 26S rDNA D1/D2 domain].Wei Sheng Wu Xue Bao. 2009 Aug;49(8):1011-7. Wei Sheng Wu Xue Bao. 2009. PMID: 19835161 Chinese.
-
Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United kingdom mycology reference laboratory.J Clin Microbiol. 2007 Apr;45(4):1152-8. doi: 10.1128/JCM.02061-06. Epub 2007 Jan 24. J Clin Microbiol. 2007. PMID: 17251397 Free PMC article.
-
Takashi Nakase's last tweet: what is the current direction of microbial taxonomy research?FEMS Yeast Res. 2019 Dec 1;19(8):foz066. doi: 10.1093/femsyr/foz066. FEMS Yeast Res. 2019. PMID: 31816016 Review.
-
A Comprehensive Review of Identification Methods for Pathogenic Yeasts: Challenges and Approaches.Adv Biomed Res. 2023 Jul 25;12:187. doi: 10.4103/abr.abr_375_22. eCollection 2023. Adv Biomed Res. 2023. PMID: 37694259 Free PMC article. Review.
Cited by
-
Fine structure of Tibetan kefir grains and their yeast distribution, diversity, and shift.PLoS One. 2014 Jun 30;9(6):e101387. doi: 10.1371/journal.pone.0101387. eCollection 2014. PLoS One. 2014. PMID: 24977409 Free PMC article.
-
D1/D2 domain of large-subunit ribosomal DNA for differentiation of Orpinomyces spp.Appl Environ Microbiol. 2011 Sep;77(18):6722-5. doi: 10.1128/AEM.05441-11. Epub 2011 Jul 22. Appl Environ Microbiol. 2011. PMID: 21784906 Free PMC article.
-
Identification, typing, antifungal resistance profile, and biofilm formation of Candida albicans isolates from Lebanese hospital patients.Biomed Res Int. 2014;2014:931372. doi: 10.1155/2014/931372. Epub 2014 Jun 1. Biomed Res Int. 2014. PMID: 24982915 Free PMC article.
-
Molecular analysis and anticancer properties of two identified isolates, Fusarium solani and Emericella nidulans isolated from Wady El-Natron soil in Egypt against Caco-2 (ATCC) cell line.Asian Pac J Trop Biomed. 2012 Nov;2(11):863-9. doi: 10.1016/S2221-1691(12)60244-5. Asian Pac J Trop Biomed. 2012. PMID: 23569862 Free PMC article.
-
Development of two molecular approaches for differentiation of clinically relevant yeast species closely related to Candida guilliermondii and Candida famata.J Clin Microbiol. 2014 Sep;52(9):3190-5. doi: 10.1128/JCM.01297-14. Epub 2014 Jun 20. J Clin Microbiol. 2014. PMID: 24951804 Free PMC article.
References
-
- Alonso-Vargas, R., L. Elorduy, E. Eraso, F. J. Cano, J. Guarro, J. Ponton, and G. Quindos. 2008. Isolation of Candida africana, probable atypical strains of Candida albicans, from a patient with vaginitis. Med. Mycol. 46:167-170. - PubMed
-
- Brito, E. H., R. S. Brilhante, R. A. Cordeiro, J. J. Sidrim, R. O. Fontenelle, L. M. Melo, E. S. Albuquerque, and M. F. Rocha. 2009. PCR-AGE, automated and manual methods to identify Candida strains from veterinary sources: a comparative approach. Vet. Microbiol. 139:318-322. - PubMed
-
- Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. LaFe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

