Epidemiologic study of human influenza virus infection in South Korea from 1999 to 2007: origin and evolution of A/Fujian/411/2002-like strains
- PMID: 20392920
- PMCID: PMC2884524
- DOI: 10.1128/JCM.00209-10
Epidemiologic study of human influenza virus infection in South Korea from 1999 to 2007: origin and evolution of A/Fujian/411/2002-like strains
Abstract
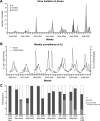
Influenza epidemics arise through the accumulation of viral genetic changes, culminating in a novel antigenic type that is able to escape host immunity. Following an outbreak of the A/Fujian/411/2002-like strains in Asia, including China, Japan, and South Korea, in 2002, Australia and New Zealand experienced substantial outbreaks of the same strains in 2003, and subsequently worldwide outbreaks occurred in the 2003-2004 season. The emergence of A/Fujian/411/2002-like strains coincided with a higher level of influenza-like illness in South Korea than what is seen at the peak of a normal season, and there was at least a year's difference between South Korea and the United States. Genetic evolution of human influenza A/H3N2 viruses was monitored by sequence analysis of hemagglutinin (HA) genes collected in Asia, including 269 (164 new) HA genes isolated in South Korea from 1999 to 2007. The Fujian-like influenza strains were disseminated with rapid sequence variation across the antigenic sites of the HA1 domain, which sharply distinguished between the A/Moscow/10/1999-like and A/Fujian/411/2002-like strains. This fast variation, equivalent to approximately 10 amino acid changes within a year, occurred in Asia and would be the main cause of the disappearance of the reassortants, although the reassortant and nonreassortant Fujian-like strains circulated simultaneously in Asia.
Figures



References
-
- Anisimova, M., and O. Gascuel. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539-552. - PubMed
-
- Barr, I. G., N. Komadina, A. C. Hurt, P. Iannello, C. Tomasov, R. Shaw, C. Durrant, H. Sjogren, and A. W. Hampson. 2005. An influenza A(H3) reassortant was epidemic in Australia and New Zealand in 2003. J. Med. Virol. 76:391-397. - PubMed
-
- Besselaar, T. G., L. Botha, J. M. McAnerney, and B. D. Schoub. 2004. Antigenic and molecular analysis of influenza A (H3N2) virus strains isolated from a localised influenza outbreak in South Africa in 2003. J. Med. Virol. 73:71-78. - PubMed
-
- Bush, R. M., C. A. Bender, K. Subbarao, N. J. Cox, and W. M. Fitch. 1999. Predicting the evolution of human influenza A. Science 286:1921-1925. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials

