Widespread transcription at neuronal activity-regulated enhancers
- PMID: 20393465
- PMCID: PMC3020079
- DOI: 10.1038/nature09033
Widespread transcription at neuronal activity-regulated enhancers
Abstract
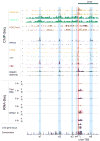
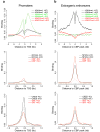
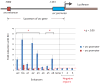
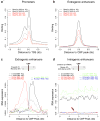
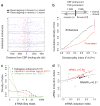
We used genome-wide sequencing methods to study stimulus-dependent enhancer function in mouse cortical neurons. We identified approximately 12,000 neuronal activity-regulated enhancers that are bound by the general transcriptional co-activator CBP in an activity-dependent manner. A function of CBP at enhancers may be to recruit RNA polymerase II (RNAPII), as we also observed activity-regulated RNAPII binding to thousands of enhancers. Notably, RNAPII at enhancers transcribes bi-directionally a novel class of enhancer RNAs (eRNAs) within enhancer domains defined by the presence of histone H3 monomethylated at lysine 4. The level of eRNA expression at neuronal enhancers positively correlates with the level of messenger RNA synthesis at nearby genes, suggesting that eRNA synthesis occurs specifically at enhancers that are actively engaged in promoting mRNA synthesis. These findings reveal that a widespread mechanism of enhancer activation involves RNAPII binding and eRNA synthesis.
Figures
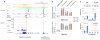
Comment in
-
Transcription: Enhancers make non-coding RNA.Nature. 2010 May 13;465(7295):173-4. doi: 10.1038/465173a. Nature. 2010. PMID: 20463730 No abstract available.
References
-
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27(2 Pt 1):299–308. - PubMed
-
- Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59(6):846–860. - PubMed
-
- Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

