Ectopic expression of wild-type FGFR3 cooperates with MYC to accelerate development of B-cell lineage neoplasms
- PMID: 20393505
- PMCID: PMC3118571
- DOI: 10.1038/leu.2010.50
Ectopic expression of wild-type FGFR3 cooperates with MYC to accelerate development of B-cell lineage neoplasms
Abstract
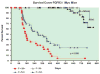
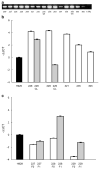
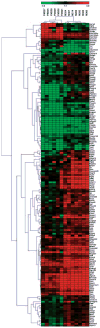
The t(4;14) translocation in multiple myeloma (MM) simultaneously dysregulates two apparent oncogenes: fibroblast growth factor receptor 3 (FGFR3) controlled by the 3' immunoglobulin heavy chain enhancer on der(14) and MMSET controlled by the intronic Emu enhancer on der(4). Although all MM tumors and cell lines with a t(4;14) translocation have dysregulated MMSET, about 25% do not express FGFR3. Therefore, the function of dysregulated wild-type (WT) FGFR3 in the pathogenesis of MM remains unclear. We developed a murine transgenic (TG) model in which WT FGFR3 is overexpressed in B lymphoid cells. Although high levels of FGFR3 resulted in lymphoid hyperplasia in about one-third of older mice, no increase in tumorigenesis was observed. However, double TG FGFR3/Myc mice develop mature B lymphoma tumors that occur with a higher penetrance and shorter latency than in single TG Myc mice (P=0.006). We conclude that expression of high levels of WT FGFR3 can be oncogenic and cooperate with MYC to generate B lymphoid tumors. This suggests that dysregulated FGFR3 expression is likely to be essential at least for the early stages of pathogenesis of MM tumors that have a t(4;14) translocation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

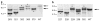
References
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. - PubMed
-
- Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2:175–187. - PubMed
-
- Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–6338. - PubMed
-
- Fonseca R, Blood E, Rue M, Harrington D, Oken MM, Kyle RA, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–4575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

