Real-time tRNA transit on single translating ribosomes at codon resolution
- PMID: 20393556
- PMCID: PMC4466108
- DOI: 10.1038/nature08925
Real-time tRNA transit on single translating ribosomes at codon resolution
Abstract
Translation by the ribosome occurs by a complex mechanism involving the coordinated interaction of multiple nucleic acid and protein ligands. Here we use zero-mode waveguides (ZMWs) and sophisticated detection instrumentation to allow real-time observation of translation at physiologically relevant micromolar ligand concentrations. Translation at each codon is monitored by stable binding of transfer RNAs (tRNAs)-labelled with distinct fluorophores-to translating ribosomes, which allows direct detection of the identity of tRNA molecules bound to the ribosome and therefore the underlying messenger RNA (mRNA) sequence. We observe the transit of tRNAs on single translating ribosomes and determine the number of tRNA molecules simultaneously bound to the ribosome, at each codon of an mRNA molecule. Our results show that ribosomes are only briefly occupied by two tRNA molecules and that release of deacylated tRNA from the exit (E) site is uncoupled from binding of aminoacyl-tRNA site (A-site) tRNA and occurs rapidly after translocation. The methods outlined here have broad application to the study of mRNA sequences, and the mechanism and regulation of translation.
Figures

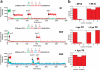
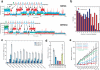

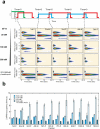
Comment in
-
Single-molecule analysis: A ribosome in action.Nature. 2010 Apr 15;464(7291):987-8. doi: 10.1038/464987a. Nature. 2010. PMID: 20393548 No abstract available.
-
A glow on protein synthesis.Nat Methods. 2010 Jun;7(6):422-3. doi: 10.1038/nmeth0610-422a. Nat Methods. 2010. PMID: 20521368
References
-
- Green R, Noller HF. Ribosomes and translation. Annu. Rev. Biochem. 1997;66:679–716. - PubMed
-
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. - PubMed
-
- Hausner TP, Geigenmüller U, Nierhaus KH. The allosteric three –site model for the ribosomal elongation cycle. New insights into the inhibition mechanism of aminoglycosides, thiostrepton, and viomycin. J. Biol. Chem. 1988;263:13103–13111. - PubMed
-
- Rodnina MW, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome : kinetic and structural mechanisms. Annu. Rev. Biochem. 2001;70:415–435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

