Dissection of genetically complex traits with extremely large pools of yeast segregants
- PMID: 20393561
- PMCID: PMC2862354
- DOI: 10.1038/nature08923
Dissection of genetically complex traits with extremely large pools of yeast segregants
Abstract
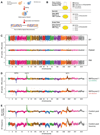
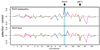
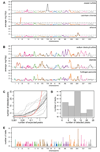
Most heritable traits, including many human diseases, are caused by multiple loci. Studies in both humans and model organisms, such as yeast, have failed to detect a large fraction of the loci that underlie such complex traits. A lack of statistical power to identify multiple loci with small effects is undoubtedly one of the primary reasons for this problem. We have developed a method in yeast that allows the use of much larger sample sizes than previously possible and hence permits the detection of multiple loci with small effects. The method involves generating very large numbers of progeny from a cross between two Saccharomyces cerevisiae strains and then phenotyping and genotyping pools of these offspring. We applied the method to 17 chemical resistance traits and mitochondrial function, and identified loci for each of these phenotypes. We show that the level of genetic complexity underlying these quantitative traits is highly variable, with some traits influenced by one major locus and others by at least 20 loci. Our results provide an empirical demonstration of the genetic complexity of a number of traits and show that it is possible to identify many of the underlying factors using straightforward techniques. Our method should have broad applications in yeast and can be extended to other organisms.
Figures

Comment in
-
Genomics: Lessons in complexity from yeast.Nature. 2010 Apr 15;464(7291):985-6. doi: 10.1038/464985a. Nature. 2010. PMID: 20393546 No abstract available.
-
Genomic dark matter: the search goes extreme.Nat Methods. 2010 Jun;7(6):424. doi: 10.1038/nmeth0610-424. Nat Methods. 2010. PMID: 20524216
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

