CpG islands influence chromatin structure via the CpG-binding protein Cfp1
- PMID: 20393567
- PMCID: PMC3730110
- DOI: 10.1038/nature08924
CpG islands influence chromatin structure via the CpG-binding protein Cfp1
Abstract
CpG islands (CGIs) are prominent in the mammalian genome owing to their GC-rich base composition and high density of CpG dinucleotides. Most human gene promoters are embedded within CGIs that lack DNA methylation and coincide with sites of histone H3 lysine 4 trimethylation (H3K4me3), irrespective of transcriptional activity. In spite of these intriguing correlations, the functional significance of non-methylated CGI sequences with respect to chromatin structure and transcription is unknown. By performing a search for proteins that are common to all CGIs, here we show high enrichment for Cfp1, which selectively binds to non-methylated CpGs in vitro. Chromatin immunoprecipitation of a mono-allelically methylated CGI confirmed that Cfp1 specifically associates with non-methylated CpG sites in vivo. High throughput sequencing of Cfp1-bound chromatin identified a notable concordance with non-methylated CGIs and sites of H3K4me3 in the mouse brain. Levels of H3K4me3 at CGIs were markedly reduced in Cfp1-depleted cells, consistent with the finding that Cfp1 associates with the H3K4 methyltransferase Setd1 (refs 7, 8). To test whether non-methylated CpG-dense sequences are sufficient to establish domains of H3K4me3, we analysed artificial CpG clusters that were integrated into the mouse genome. Despite the absence of promoters, the insertions recruited Cfp1 and created new peaks of H3K4me3. The data indicate that a primary function of non-methylated CGIs is to genetically influence the local chromatin modification state by interaction with Cfp1 and perhaps other CpG-binding proteins.
Figures
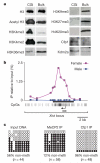


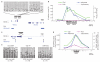
References
-
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. - PubMed
-
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature Rev. Genet. 2008;9:465–476. - PubMed
-
- Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Lee JH, Skalnik DG. CpG-binding protein is a nuclear matrix- and euchromatin-associated protein localized to nuclear speckles containing human trithorax. Identification of nuclear matrix targeting signals. J. Biol. Chem. 2002;277:42259–42267. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

