PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila
- PMID: 20394051
- PMCID: PMC2881544
- DOI: 10.1002/cne.22311
PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila
Abstract
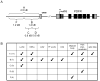
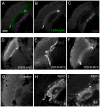
Daily rhythms of behavior are controlled by a circuit of circadian pacemaking neurons. In Drosophila, 150 pacemakers participate in this network, and recent observations suggest that the network is divisible into M and E oscillators, which normally interact and synchronize. Sixteen oscillator neurons (the small and large lateral neurons [LNvs]) express a neuropeptide called pigment-dispersing factor (PDF) whose signaling is often equated with M oscillator output. Given the significance of PDF signaling to numerous aspects of behavioral and molecular rhythms, determining precisely where and how signaling via the PDF receptor (PDFR) occurs is now a central question in the field. Here we show that GAL4-mediated rescue of pdfr phenotypes using a UAS-PDFR transgene is insufficient to provide complete behavioral rescue. In contrast, we describe a approximately 70-kB PDF receptor (pdfr) transgene that does rescue the entire pdfr circadian behavioral phenotype. The transgene is widely but heterogeneously expressed among pacemakers, and also among a limited number of non-pacemakers. Our results support an important hypothesis: the small LNv cells directly target a subset of the other crucial pacemaker neurons cells. Furthermore, expression of the transgene confirms an autocrine feedback signaling by PDF back to PDF-expressing cells. Finally, the results present an unexpected PDF receptor site: the large LNv cells appear to target a population of non-neuronal cells that resides at the base of the eye.
(c) 2009 Wiley-Liss, Inc.
Figures



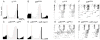



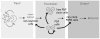
Similar articles
-
The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior.PLoS Biol. 2009 Jul;7(7):e1000154. doi: 10.1371/journal.pbio.1000154. Epub 2009 Jul 21. PLoS Biol. 2009. PMID: 19621061 Free PMC article.
-
Functional PDF Signaling in the Drosophila Circadian Neural Circuit Is Gated by Ral A-Dependent Modulation.Neuron. 2016 May 18;90(4):781-794. doi: 10.1016/j.neuron.2016.04.002. Epub 2016 May 5. Neuron. 2016. PMID: 27161526 Free PMC article.
-
Drosophila DH31 Neuropeptide and PDF Receptor Regulate Night-Onset Temperature Preference.J Neurosci. 2016 Nov 16;36(46):11739-11754. doi: 10.1523/JNEUROSCI.0964-16.2016. J Neurosci. 2016. PMID: 27852781 Free PMC article.
-
Mechanisms of clock output in the Drosophila circadian pacemaker system.J Biol Rhythms. 2006 Dec;21(6):445-57. doi: 10.1177/0748730406293910. J Biol Rhythms. 2006. PMID: 17107935 Review.
-
Circadian pathway: the other shoe drops.Curr Biol. 2005 Dec 20;15(24):R987-9. doi: 10.1016/j.cub.2005.11.053. Curr Biol. 2005. PMID: 16360675 Review.
Cited by
-
Uncovering the Roles of Clocks and Neural Transmission in the Resilience of Drosophila Circadian Network.Front Physiol. 2021 May 26;12:663339. doi: 10.3389/fphys.2021.663339. eCollection 2021. Front Physiol. 2021. PMID: 34122135 Free PMC article.
-
GW182 controls Drosophila circadian behavior and PDF-receptor signaling.Neuron. 2013 Apr 10;78(1):152-65. doi: 10.1016/j.neuron.2013.01.035. Neuron. 2013. PMID: 23583112 Free PMC article.
-
Seasonal cues act through the circadian clock and pigment-dispersing factor to control EYES ABSENT and downstream physiological changes.Curr Biol. 2023 Feb 27;33(4):675-687.e5. doi: 10.1016/j.cub.2023.01.006. Epub 2023 Jan 27. Curr Biol. 2023. PMID: 36708710 Free PMC article.
-
Wakefulness Is Promoted during Day Time by PDFR Signalling to Dopaminergic Neurons in Drosophila melanogaster.eNeuro. 2018 Aug 8;5(4):ENEURO.0129-18.2018. doi: 10.1523/ENEURO.0129-18.2018. eCollection 2018 Jul-Aug. eNeuro. 2018. PMID: 30131970 Free PMC article.
-
The Neuronal Circuit of the Dorsal Circadian Clock Neurons in Drosophila melanogaster.Front Physiol. 2022 Apr 29;13:886432. doi: 10.3389/fphys.2022.886432. eCollection 2022. Front Physiol. 2022. PMID: 35574472 Free PMC article.
References
-
- Alfonso TB, Jones BW. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev Biol. 2002;248:369–383. - PubMed
-
- Baker DA, Beckingham KM, Armstrong JD. Functional dissection of the neural substrates for gravitaxic maze behavior in Drosophila melanogaster. J Comp Neurol. 2007;501:756–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

