Frontal affinity chromatography-mass spectrometry useful for characterization of new ligands for GPR17 receptor
- PMID: 20394377
- PMCID: PMC6201305
- DOI: 10.1021/jm901691y
Frontal affinity chromatography-mass spectrometry useful for characterization of new ligands for GPR17 receptor
Abstract
The application of frontal affinity chromatography-mass spectrometry (FAC-MS), along with molecular modeling studies, to the screening of potential drug candidates toward the recently deorphanized G-protein-coupled receptor (GPCR) GPR17 is shown. GPR17 is dually activated by uracil nucleotides and cysteinyl-leukotrienes, and is expressed in organs typically undergoing ischemic damage (i.e., brain, heart and kidney), thus representing a new pharmacological target for acute and chronic neurodegeneration. GPR17 was entrapped on an immobilized artificial membrane (IAM), and this stationary phase was used to screen a library of nucleotide derivatives by FAC-MS to select high affinity ligands. The chromatographic results have been validated with a reference functional assay ([(35)S]GTPgammaS binding assay). The receptor nucleotide-binding site was studied by setting up a column where a mutated GPR17 receptor (Arg255Ile) has been immobilized. The chromatographic behavior of the tested nucleotide derivatives together with in silico studies have been used to gain insights into the structure requirement of GPR17 ligands.
Figures
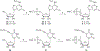


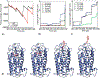

Similar articles
-
Synthesis and Ability of New Ligands for G Protein-Coupled Receptors 17 (GPR17).Med Sci Monit. 2017 Feb 22;23:953-959. doi: 10.12659/msm.902048. Med Sci Monit. 2017. PMID: 28223679 Free PMC article.
-
Development of an immobilized GPR17 receptor stationary phase for binding determination using frontal affinity chromatography coupled to mass spectrometry.Anal Biochem. 2009 Jan 1;384(1):123-9. doi: 10.1016/j.ab.2008.09.010. Epub 2008 Sep 15. Anal Biochem. 2009. PMID: 18835238
-
The orphan receptor GPR17 identified as a new dual uracil nucleotides/cysteinyl-leukotrienes receptor.EMBO J. 2006 Oct 4;25(19):4615-27. doi: 10.1038/sj.emboj.7601341. Epub 2006 Sep 21. EMBO J. 2006. PMID: 16990797 Free PMC article.
-
The G Protein-Coupled Receptor GPR17: Overview and Update.ChemMedChem. 2016 Dec 6;11(23):2567-2574. doi: 10.1002/cmdc.201600453. Epub 2016 Nov 14. ChemMedChem. 2016. PMID: 27863043 Review.
-
GPR17 receptor modulators and their therapeutic implications: review of recent patents.Expert Opin Ther Pat. 2019 Feb;29(2):85-95. doi: 10.1080/13543776.2019.1568990. Epub 2019 Jan 24. Expert Opin Ther Pat. 2019. PMID: 30640576 Review.
Cited by
-
Oscillatory calcium release and sustained store-operated oscillatory calcium signaling prevents differentiation of human oligodendrocyte progenitor cells.Sci Rep. 2022 Apr 13;12(1):6160. doi: 10.1038/s41598-022-10095-1. Sci Rep. 2022. PMID: 35418597 Free PMC article.
-
Phenotypic changes, signaling pathway, and functional correlates of GPR17-expressing neural precursor cells during oligodendrocyte differentiation.J Biol Chem. 2011 Mar 25;286(12):10593-604. doi: 10.1074/jbc.M110.162867. Epub 2011 Jan 5. J Biol Chem. 2011. PMID: 21209081 Free PMC article.
-
Solid-Supported Proteins in the Liquid Chromatography Domain to Probe Ligand-Target Interactions.Front Chem. 2019 Nov 15;7:752. doi: 10.3389/fchem.2019.00752. eCollection 2019. Front Chem. 2019. PMID: 31803714 Free PMC article. Review.
-
Altered Purinergic Signaling in Neurodevelopmental Disorders: Focus on P2 Receptors.Biomolecules. 2023 May 18;13(5):856. doi: 10.3390/biom13050856. Biomolecules. 2023. PMID: 37238724 Free PMC article. Review.
-
Synthesis and Ability of New Ligands for G Protein-Coupled Receptors 17 (GPR17).Med Sci Monit. 2017 Feb 22;23:953-959. doi: 10.12659/msm.902048. Med Sci Monit. 2017. PMID: 28223679 Free PMC article.
References
-
- Schriemer DC; Bundle DR; Li L; Hindsgaul O Micro-scale frontal affinity chromatography with mass spectrometric detection: a new method for the screening of compound libraries. Angew. Chem 1998,110,3625.-; Angew Chem.,Int.Ed.1998,37,3383–3387. - PubMed
-
- (Ng E; Schriemer D Emerging challenges in ligand discovery: new opportunities for chromatographic assay. Expert Rev. Proteomics 2005,2,891.-. - PubMed
-
- Called E; Temporini C; Caccialanza G; Massolini G Target- based drug discovery: the emerging success of frontal affinity chromatography coupled to mass spectrometry. ChemMedChem 2009,4,905.-. - PubMed
-
- Slon-Usakiewicz JJ; Dai JR; Ng W; Foster JE; Deretey E; Toledo-Sherman L; Redden PR; Pasternak A; Reid N Kinase screening. Applications of frontal affinity chromatography coupled to mass spectrometry in drug discovery. Anal.Chem. 2005,77,1268–1274. - PubMed
-
- Ng ES; Chan NW; Lewis DF; Hindsgaul O; Schriemer DC Frontal affinity chromatography−mass spectrometry. Nat.Protoc 2007,2,1907.-. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

