Molecular diversity through RNA editing: a balancing act
- PMID: 20395010
- PMCID: PMC2865426
- DOI: 10.1016/j.tig.2010.02.001
Molecular diversity through RNA editing: a balancing act
Abstract
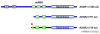
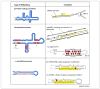
RNA editing by adenosine deamination fuels the generation of RNA and protein diversity in eukaryotes, particularly in higher organisms. This includes the recoding of translated exons, widespread editing of retrotransposon-derived repeat elements and sequence modification of microRNA (miRNA) transcripts. Such changes can bring about specific amino acid substitutions, alternative splicing and changes in gene expression levels. Although the overall prevalence of adenosine-to-inosine (A-to-I) editing and its specific functional impact on many of the affected genes is not yet known, the importance of balancing RNA modification levels across time and space is becoming increasingly evident. In particular, transcriptome instabilities in the form of too much or too little RNA editing activity, or misguided editing, manifest in several human disease phenotypes and can disrupt that balance.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
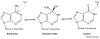

References
-
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. - PubMed
-
- Gommans WM, et al. Diversifying Exon Code through A-to-I RNA Editing. In: Smith H, editor. DNA RNA Editing. Wiley & Sons, Inc; 2008. pp. 3–30.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

