Rapid diversification of cell signaling phenotypes by modular domain recombination
- PMID: 20395511
- PMCID: PMC2975375
- DOI: 10.1126/science.1182376
Rapid diversification of cell signaling phenotypes by modular domain recombination
Abstract
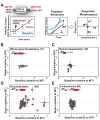
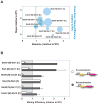
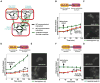
Cell signaling proteins are often modular, containing distinct catalytic and regulatory domains. Recombination of such biological modules has been proposed to be a major source of evolutionary innovation. We systematically analyzed the phenotypic diversity of a signaling response that results from domain recombination by using 11 proteins in the yeast mating pathway to construct a library of 66 chimeric domain recombinants. Domain recombination resulted in greater diversity in pathway response dynamics than did duplication of genes, of single domains, or of two unlinked domains. Domain recombination also led to changes in mating phenotype, including recombinants with increased mating efficiency over the wild type. Thus, novel linkages between preexisting domains may have a major role in the evolution of protein networks and novel phenotypic behaviors.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
Signalling: Making new connections.Nat Rev Genet. 2010 Jun;11(6):387. doi: 10.1038/nrg2801. Epub 2010 May 11. Nat Rev Genet. 2010. PMID: 20458345 No abstract available.
-
Cell signalling: Making new connections.Nat Rev Mol Cell Biol. 2010 Jun;11(6):386. doi: 10.1038/nrm2909. Epub 2010 May 12. Nat Rev Mol Cell Biol. 2010. PMID: 20461096 No abstract available.
References
-
- Chothia C, Gough J, Vogel C, Teichmann SA. Science. 2003 Jun 13;300:1701. - PubMed
-
- Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Curr Opin Struct Biol. 2004 Apr;14:208. - PubMed
-
- Pawson T, Nash P. Science. 2003 Apr 18;300:445. - PubMed
-
- Voigt CA, Martinez C, Wang ZG, Mayo SL, Arnold FH. Nat Struct Biol. 2002 Jul;9:553. - PubMed
-
- Bashton M, Chothia C. Structure. 2007 Jan;15:85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

