Identification of RanBP 9/10 as interacting partners for protein kinase C (PKC) gamma/delta and the D1 dopamine receptor: regulation of PKC-mediated receptor phosphorylation
- PMID: 20395553
- PMCID: PMC2912060
- DOI: 10.1124/mol.110.063727
Identification of RanBP 9/10 as interacting partners for protein kinase C (PKC) gamma/delta and the D1 dopamine receptor: regulation of PKC-mediated receptor phosphorylation
Abstract
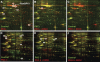
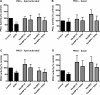
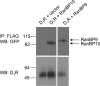
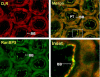
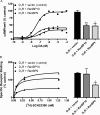
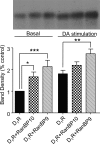
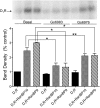
We reported previously that ethanol treatment regulates D(1) receptor phosphorylation and signaling in a protein kinase C (PKC) delta- and PKCgamma-dependent fashion by a mechanism that may involve PKC isozyme-specific interacting proteins. Using a PKC isozyme-specific coimmunoprecipitation approach coupled to mass spectrometry, we report the identification of RanBP9 and RanBP10 as novel interacting proteins for both PKCgamma and PKCdelta. Both RanBP9 and RanBP10 were found to specifically coimmunoprecipitate with both PKCgamma and PKCdelta; however, this association did not seem to mediate the ethanol regulation of the PKCs. It is noteworthy that the D(1) receptor was also found to specifically coimmunoprecipitate with RanBP9/10 from human embryonic kidney (HEK) 293T cells and with endogenous RanBP9 from rat kidney. RanBP9 and RanBP10 were also found to colocalize at the cellular level with the D(1) receptor in both kidney and brain tissue. Although overexpression of RanBP9 or RanBP10 in HEK293T cells did not seem to alter the kinase activities of either PKCdelta or PKCgamma, both RanBP proteins regulated D(1) receptor phosphorylation, signaling, and, in the case of RanBP9, expression. Specifically, overexpression of either RanBP9 or RanBP10 enhanced basal D(1) receptor phosphorylation, which was associated with attenuation of D(1) receptor-stimulated cAMP accumulation. Moreover, treatment of cells with select PKC inhibitors blocked the RanBP9/10-dependent increase in basal receptor phosphorylation, suggesting that phosphorylation of the receptor by PKC is regulated by RanBP9/10. These data support the idea that RanBP9 and RanBP10 may function as signaling integrators and dictate the efficient regulation of D(1) receptor signaling by PKCdelta and PKCgamma.
Figures
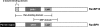
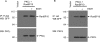


References
-
- Bermak JC, Li M, Bullock C, Zhou QY. (2001) Regulation of transport of the dopamine D1 receptor by a new membrane-associated ER protein. Nat Cell Biol 3:492–498 - PubMed
-
- Besson A, Wilson TL, Yong VW. (2002) The anchoring protein RACK1 links protein kinase Cepsilon to integrin beta chains. Requirements for adhesion and motility. J Biol Chem 277:22073–22084 - PubMed
-
- Csukai M, Chen CH, De Matteis MA, Mochly-Rosen D. (1997) The coatomer protein beta'-COP, a selective binding protein (RACK) for protein kinase Cepsilon. J Biol Chem 272:29200–29206 - PubMed
-
- Denti S, Sirri A, Cheli A, Rogge L, Innamorati G, Putignano S, Fabbri M, Pardi R, Bianchi E. (2004) RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J Biol Chem 279:13027–13034 - PubMed
-
- D'Souza MS, Ikegami A, Olsen CM, Duvauchelle CL. (2003) Chronic D1 agonist and ethanol coadministration facilitate ethanol-mediated behaviors. Pharmacol Biochem Behav 76:335–342 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

