Genome-wide association studies in diverse populations
- PMID: 20395969
- PMCID: PMC3079573
- DOI: 10.1038/nrg2760
Genome-wide association studies in diverse populations
Abstract
Genome-wide association (GWA) studies have identified a large number of SNPs associated with disease phenotypes. As most GWA studies have been performed in populations of European descent, this Review examines the issues involved in extending the consideration of GWA studies to diverse worldwide populations. Although challenges exist with issues such as imputation, admixture and replication, investigation of a greater diversity of populations could make substantial contributions to the goal of mapping the genetic determinants of complex diseases for the human population as a whole.
Figures
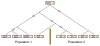
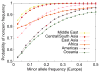
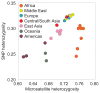
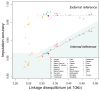
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

