CenH3/CID incorporation is not dependent on the chromatin assembly factor CHD1 in Drosophila
- PMID: 20396651
- PMCID: PMC2852906
- DOI: 10.1371/journal.pone.0010120
CenH3/CID incorporation is not dependent on the chromatin assembly factor CHD1 in Drosophila
Abstract
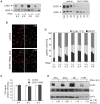
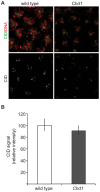
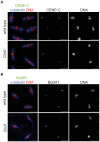
CHD1 is a SNF2-related ATPase that is required for the genome-wide incorporation of variant histone H3.3 in the paternal pronucleus as well as in transcriptionally active nuclei in Drosophila embryos. The S. pombe and vertebrate orthologs of CHD1 have been implicated in the assembly of the centromeric histone H3 variant CenH3(CENP-A), which occurs in a DNA replication-independent manner. Here, we examined whether CHD1 participates in the assembly of CenH3(CID) in Drosophila. In contrast to the findings in fission yeast and vertebrate cells, our evidence clearly argues against such a role for CHD1 in Drosophila. CHD1 does not localize to centromeres in either S2 cells or developing fly embryos. Down-regulation of CHD1 in S2 cells by RNAi reveals unchanged levels of CenH3(CID) at the centromeres. Most notably, ablation of functional CHD1 in Chd1 mutant fly embryos does not interfere with centromere and kinetochore assembly, as the levels and localization of CenH3(CID), CENP-C and BubR1 in the mutant embryos remain similar to those seen in wild-type embryos. These results indicate that Drosophila CHD1 has no direct function in the incorporation of the centromeric H3 variant CenH3(CID) into chromatin. Therefore, centromeric chromatin assembly may involve different mechanisms in different organisms.
Conflict of interest statement
Figures



References
-
- Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Curr Opin Genet Dev. 2005;15:177–184. - PubMed
-
- Haushalter KA, Kadonaga JT. Chromatin assembly by DNA-translocating motors. Nat Rev Mol Cell Biol. 2003;4:613–620. - PubMed
-
- Polo SE, Almouzni G. Chromatin assembly: a basic recipe with various flavours. Curr Opin Genet Dev. 2006;16:104–111. - PubMed
-
- Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol. 2005;12:160–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

