Elongator function in tRNA wobble uridine modification is conserved between yeast and plants
- PMID: 20398216
- PMCID: PMC2904499
- DOI: 10.1111/j.1365-2958.2010.07163.x
Elongator function in tRNA wobble uridine modification is conserved between yeast and plants
Erratum in
- Mol Microbiol. 2010 Jul;77(2):531
Abstract
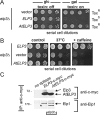
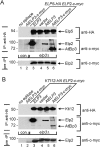
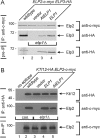
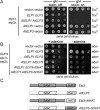
Based on studies in yeast and mammalian cells the Elongator complex has been implicated in functions as diverse as histone acetylation, polarized protein trafficking and tRNA modification. Here we show that Arabidopsis mutants lacking the Elongator subunit AtELP3/ELO3 have a defect in tRNA wobble uridine modification. Moreover, we demonstrate that yeast elp3 and elp1 mutants expressing the respective Arabidopsis Elongator homologues AtELP3/ELO3 and AtELP1/ELO2 assemble integer Elongator complexes indicating a high degree of structural conservation. Surprisingly, in vivo complementation studies based on Elongator-dependent tRNA nonsense suppression and zymocin tRNase toxin assays indicated that while AtELP1 rescued defects of a yeast elp1 mutant, the most conserved Elongator gene AtELP3, failed to complement an elp3 mutant. This lack of complementation is due to incompatibility with yeast ELP1 as coexpression of both plant genes in an elp1 elp3 yeast mutant restored Elongator's tRNA modification function in vivo. Similarly, AtELP1, not ScELP1 also supported partial complementation by yeast-plant Elp3 hybrids suggesting that AtElp1 has less stringent sequence requirements for Elp3 than ScElp1. We conclude that yeast and plant Elongator share tRNA modification roles and propose that this function might be conserved in Elongator from all eukaryotic kingdoms of life.
Figures

Similar articles
-
Phosphorylation of Elp1 by Hrr25 is required for elongator-dependent tRNA modification in yeast.PLoS Genet. 2015 Jan 8;11(1):e1004931. doi: 10.1371/journal.pgen.1004931. eCollection 2015 Jan. PLoS Genet. 2015. PMID: 25569479 Free PMC article.
-
A conserved and essential basic region mediates tRNA binding to the Elp1 subunit of the Saccharomyces cerevisiae Elongator complex.Mol Microbiol. 2014 Jun;92(6):1227-42. doi: 10.1111/mmi.12624. Epub 2014 May 19. Mol Microbiol. 2014. PMID: 24750273 Free PMC article.
-
Use of a Yeast tRNase Killer Toxin to Diagnose Kti12 Motifs Required for tRNA Modification by Elongator.Toxins (Basel). 2017 Sep 5;9(9):272. doi: 10.3390/toxins9090272. Toxins (Basel). 2017. PMID: 28872616 Free PMC article.
-
Elongator, a conserved complex required for wobble uridine modifications in eukaryotes.RNA Biol. 2014;11(12):1519-28. doi: 10.4161/15476286.2014.992276. RNA Biol. 2014. PMID: 25607684 Free PMC article. Review.
-
How Elongator Acetylates tRNA Bases.Int J Mol Sci. 2020 Nov 3;21(21):8209. doi: 10.3390/ijms21218209. Int J Mol Sci. 2020. PMID: 33152999 Free PMC article. Review.
Cited by
-
Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi.Nat Struct Mol Biol. 2016 Sep;23(9):794-802. doi: 10.1038/nsmb.3265. Epub 2016 Jul 25. Nat Struct Mol Biol. 2016. PMID: 27455459 Free PMC article.
-
Translational control of cell division by Elongator.Cell Rep. 2012 May 31;1(5):424-33. doi: 10.1016/j.celrep.2012.04.001. Cell Rep. 2012. PMID: 22768388 Free PMC article.
-
DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator.J Biol Chem. 2012 Sep 21;287(39):32535-45. doi: 10.1074/jbc.M112.402727. Epub 2012 Aug 1. J Biol Chem. 2012. PMID: 22854966 Free PMC article.
-
AtELP4 a subunit of the Elongator complex in Arabidopsis, mediates cell proliferation and dorsoventral polarity during leaf morphogenesis.Front Plant Sci. 2022 Oct 21;13:1033358. doi: 10.3389/fpls.2022.1033358. eCollection 2022. Front Plant Sci. 2022. PMID: 36340367 Free PMC article.
-
Modify or die?--RNA modification defects in metazoans.RNA Biol. 2014;11(12):1555-67. doi: 10.4161/15476286.2014.992279. RNA Biol. 2014. PMID: 25692999 Free PMC article. Review.
References
-
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

